Relatedness of chromosomal and plasmid DNAs of Erwinia pyrifoliae and Erwinia amylovora
- PMID: 12450843
- PMCID: PMC134437
- DOI: 10.1128/AEM.68.12.6182-6192.2002
Relatedness of chromosomal and plasmid DNAs of Erwinia pyrifoliae and Erwinia amylovora
Abstract
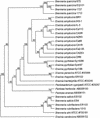
The plant pathogen Erwinia pyrifoliae has been classified as a separate species from Erwinia amylovora based in part on differences in molecular properties. In this study, these and other molecular properties were examined for E. pyrifoliae and for additional strains of E. amylovora, including strains from brambles (Rubus spp.). The nucleotide composition of the internal transcribed spacer (ITS) region was determined for six of the seven 16S-23S rRNA operons detected in these species with a 16S rRNA gene probe. Each species contained four operons with a tRNA(Glu) gene and two with tRNA(Ile) and tRNA(Ala) genes, and analysis of the operons from five strains of E. amylovora indicated a high degree of ITS variability among them. One tRNA(Glu)-containing operon from E. pyrifoliae Ep1/96 was identical to one in E. amylovora Ea110, but three tRNA(Glu) operons and two tRNA(Ile) and tRNA(Ala) operons from E. pyrifoliae contained unique nucleotide changes. When groEL sequences were used for species-specific identification, E. pyrifoliae and E. amylovora were the closest phylogenetic relatives among a set of 12 bacterial species. The placement of E. pyrifoliae distinct from E. amylovora corroborated molecular hybridization data indicating low DNA-DNA similarity between them. Determination of the nucleotide sequence of plasmid pEP36 from E. pyrifoliae Ep1/96 revealed a number of presumptive genes that matched genes previously found in pEA29 from E. amylovora and similar organization for the genes and origins of replication. Also, pEP36 and pEA29 were incompatible with clones containing the reciprocal origin regions. Finally, the ColE1-like plasmid pEP2.6 from strain Ep1/96 contained sequences found in small plasmids in E. amylovora strains IL-5 and IH3-1.
Figures

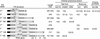
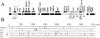
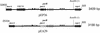
 ). Incomplete ORFs are indicated by flecked arrows (
). Incomplete ORFs are indicated by flecked arrows ( ). The position and organization of discrete families of repeats are indicated by various symbols: dotted vertical lines (¦) indicate an 8-bp repeat (ATTCTGGG) originally observed in pEP36, line arrows (→) designate a series of 3 to 14 8-bp repeats previously described in pEA29 (38). Inverted triangles (▸◂) show the position of an inverted repeat; the right repeat (◃) is incomplete in pEA29. Open arrows (➯) indicate the position of two 23-bp repeats found on pEP36 and of a single copy found on pEA29. The nine iterons are designated by solid vertical lines (|). Solid arrows (
). The position and organization of discrete families of repeats are indicated by various symbols: dotted vertical lines (¦) indicate an 8-bp repeat (ATTCTGGG) originally observed in pEP36, line arrows (→) designate a series of 3 to 14 8-bp repeats previously described in pEA29 (38). Inverted triangles (▸◂) show the position of an inverted repeat; the right repeat (◃) is incomplete in pEA29. Open arrows (➯) indicate the position of two 23-bp repeats found on pEP36 and of a single copy found on pEA29. The nine iterons are designated by solid vertical lines (|). Solid arrows ( ) indicate three 20-bp repeats found on pEP36 and on pEA29. Numbers found at either end of the drawing are the position in nucleotides of the segment depicted. The length of each segment is indicated in base pairs to the right of the drawing. An identical BamHI site is used to orient the plasmids. Maps are not to scale.
) indicate three 20-bp repeats found on pEP36 and on pEA29. Numbers found at either end of the drawing are the position in nucleotides of the segment depicted. The length of each segment is indicated in base pairs to the right of the drawing. An identical BamHI site is used to orient the plasmids. Maps are not to scale.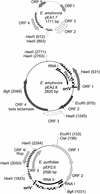
 ), respectively. The ORF coding for the RNA1 modulator protein was found on both pEP2.6 and pEA1.7 (ORF 4 on both plasmids). ORF 2 from plasmid pEA2.8 was similar to a 13.8-kDa hypothetical protein encoded on plasmid ColE1 in E. coli (accession number NP_040360). ORF 4 from pEA2.8 had 98% similarity to a beta-lactamase protein that provides resistance to ampicillin. No other ORFs had identity to any existing GenBank entries.
), respectively. The ORF coding for the RNA1 modulator protein was found on both pEP2.6 and pEA1.7 (ORF 4 on both plasmids). ORF 2 from plasmid pEA2.8 was similar to a 13.8-kDa hypothetical protein encoded on plasmid ColE1 in E. coli (accession number NP_040360). ORF 4 from pEA2.8 had 98% similarity to a beta-lactamase protein that provides resistance to ampicillin. No other ORFs had identity to any existing GenBank entries.References
-
- Chiou, C.-S., and A. L. Jones. 1991. The analysis of plasmid-mediated streptomycin resistance in Erwinia amylovora. Phytopathology 81:710-714.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

