The founder mutation MSH2*1906G-->C is an important cause of hereditary nonpolyposis colorectal cancer in the Ashkenazi Jewish population
- PMID: 12454801
- PMCID: PMC420003
- DOI: 10.1086/345075
The founder mutation MSH2*1906G-->C is an important cause of hereditary nonpolyposis colorectal cancer in the Ashkenazi Jewish population
Abstract
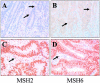
Hereditary nonpolyposis colorectal cancer (HNPCC) is caused by mutations in the mismatch-repair genes. We report here the identification and characterization of a founder mutation in MSH2 in the Ashkenazi Jewish population. We identified a nucleotide substitution, MSH2*1906G-->C, which results in a substitution of proline for alanine at codon 636 in the MSH2 protein. This allele was identified in 15 unrelated Ashkenazi Jewish families with HNPCC, most of which meet the Amsterdam criteria. Genotype analysis of 18 polymorphic loci within and flanking MSH2 suggested a single origin for the mutation. All colorectal cancers tested showed microsatellite instability and absence of MSH2 protein, by immunohistochemical analysis. In an analysis of a population-based incident series of 686 Ashkenazi Jews from Israel who have colorectal cancer, we identified 3 (0.44%) mutation carriers. Persons with a family history of colorectal or endometrial cancer were more likely to carry the mutation than were those without such a family history (P=.042), and those with colorectal cancer who carried the mutation were, on average, younger than affected individuals who did not carry it (P=.033). The mutation was not detected in either 566 unaffected Ashkenazi Jews from Israel or 1,022 control individuals from New York. In hospital-based series, the 1906C allele was identified in 5/463 Ashkenazi Jews with colorectal cancer, in 2/197 with endometrial cancer, and in 0/83 with ovarian cancer. When families identified by family history and in case series are included, 25 apparently unrelated Ashkenazi Jewish families have been found to harbor this mutation. Although this pathogenic mutation is not frequent in the Ashkenazi Jewish population (accounting for 2%-3% of colorectal cancer in those whose age at diagnosis is <60 years), it is highly penetrant and accounts for approximately one-third of HNPCC in Ashkenazi Jewish families that fulfill the Amsterdam criteria.
Figures

References
Electronic-Database Information
-
- Genome Database, The, http://gdbwww.gdb.org
-
- International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (“Choose a Mutation Database” Web page), http://www.nfdht.nl/database/mdbchoice.htm (for map information and mutations in mismatch-repair genes)
-
- National Center for Biotechnology Information “Single Nucleotide Polymorphism” Web page, http://www.ncbi.nlm.nih.gov/SNP/
-
- National Laboratory for the Genetics of Israeli Populations (Tel Aviv University), http://www.tau.ac.il/medicine/NLGIP/catalog.htm
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MSH2 [MIM 120435], MSH1 [MIM 120436], and HNPCC [MIM 114500])
References
-
- Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A, Percesepe A, Ahtola H, Harkonen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E (1998) Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med 338:1481–1487 - PubMed
-
- Andreutti-Zaugg C, Scott RJ, Iggo R (1997) Inhibition of nonsense-mediated messenger RNA decay in clinical samples facilitates detection of human MSH2 mutations with an in vivo fusion protein assay and conventional techniques. Cancer Res 57:3288–3293 - PubMed
-
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58:5248–5257 - PubMed
-
- Boyd J, Sonoda Y, Federici MG, Bogomolniy F, Rhei E, Maresco DL, Saigo PE, Almadrones LA, Barakat RR, Brown CL, Chi DS, Curtin JP, Poynor EA, Hoskins WJ (2000) Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA 283:2260–2265 - PubMed
-
- Chan TL, Yuen ST, Ho JW, Chan AS, Kwan K, Chung LP, Lam PW, Tse CW, Leung SY (2001) A novel germline 1.8-kb deletion of hMLH1 mimicking alternative splicing: a founder mutation in the Chinese population. Oncogene 20:2976–2981 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

