Aspergillus nidulans catalase-peroxidase gene (cpeA) is transcriptionally induced during sexual development through the transcription factor StuA
- PMID: 12455692
- PMCID: PMC126739
- DOI: 10.1128/EC.1.5.725-735.2002
Aspergillus nidulans catalase-peroxidase gene (cpeA) is transcriptionally induced during sexual development through the transcription factor StuA
Abstract
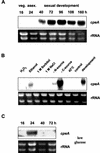
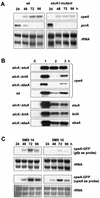
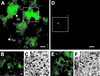
Catalases, peroxidases, and catalase-peroxidases are important enzymes to cope with reactive oxygen species in pro- and eukaryotic cells. In the filamentous fungus Aspergillus nidulans three monofunctional catalases have been described, and a fourth catalase activity was observed in native polyacrylamide gels. The latter activity is probably due to the bifunctional enzyme catalase-peroxidase, which we characterized here. The gene, named cpeA, encodes an 81-kDa polypeptide with a conserved motif for heme coordination. The enzyme comprises of two similar domains, suggesting gene duplication and fusion during evolution. The first 439 amino acids share 22% identical residues with the C terminus. Homologous proteins are found in several prokaryotes, such as Escherichia coli and Mycobacterium tuberculosis (both with 61% identity). In fungi the enzyme has been noted in Penicillium simplicissimum, Septoria tritici, and Neurospora crassa (69% identical amino acids) but is absent from Saccharomyces cerevisiae. Expression analysis in A. nidulans revealed that the gene is transcriptionally induced upon carbon starvation and during sexual development, but starvation is not sufficient to reach high levels of the transcript during development. Besides transcriptional activation, we present evidence for posttranscriptional regulation. A green fluorescent protein fusion protein localized to the cytoplasm of Hülle cells. The Hülle cell-specific expression was dependent on the developmental regulator StuA, suggesting an activating function of this helix-loop-helix transcription factor.
Figures

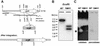



Similar articles
-
Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress.J Bacteriol. 1997 May;179(10):3284-92. doi: 10.1128/jb.179.10.3284-3292.1997. J Bacteriol. 1997. PMID: 9150225 Free PMC article.
-
Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans.Mol Microbiol. 2003 Nov;50(4):1241-55. doi: 10.1046/j.1365-2958.2003.03800.x. Mol Microbiol. 2003. PMID: 14622412
-
The Zn(II)2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture.Genetics. 2005 Feb;169(2):619-30. doi: 10.1534/genetics.104.030767. Epub 2004 Nov 1. Genetics. 2005. PMID: 15520269 Free PMC article.
-
Fungal catalases: function, phylogenetic origin and structure.Arch Biochem Biophys. 2012 Sep 15;525(2):170-80. doi: 10.1016/j.abb.2012.05.014. Epub 2012 Jun 12. Arch Biochem Biophys. 2012. PMID: 22698962 Review.
-
Evolution of structure and function of Class I peroxidases.Arch Biochem Biophys. 2010 Aug 1;500(1):45-57. doi: 10.1016/j.abb.2010.03.024. Epub 2010 Apr 4. Arch Biochem Biophys. 2010. PMID: 20371361 Review.
Cited by
-
Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts.Eukaryot Cell. 2004 Aug;3(4):835-46. doi: 10.1128/EC.3.4.835-846.2004. Eukaryot Cell. 2004. PMID: 15302816 Free PMC article. Review. No abstract available.
-
Mating type specific transcriptomic response to sex inducing pheromone in the pennate diatom Seminavis robusta.ISME J. 2021 Feb;15(2):562-576. doi: 10.1038/s41396-020-00797-7. Epub 2020 Oct 7. ISME J. 2021. PMID: 33028976 Free PMC article.
-
The APSES transcription factor LmStuA is required for sporulation, pathogenic development and effector gene expression in Leptosphaeria maculans.Mol Plant Pathol. 2015 Dec;16(9):1000-5. doi: 10.1111/mpp.12249. Epub 2015 Apr 15. Mol Plant Pathol. 2015. PMID: 25727237 Free PMC article.
-
Evaluation of catalase activity of clinical and environmental isolates of Aspergillus species.Iran J Microbiol. 2022 Feb;14(1):133-137. doi: 10.18502/ijm.v14i1.8815. Iran J Microbiol. 2022. PMID: 35664717 Free PMC article.
-
Use of laccase as a novel, versatile reporter system in filamentous fungi.Appl Environ Microbiol. 2006 Jul;72(7):5020-6. doi: 10.1128/AEM.00060-06. Appl Environ Microbiol. 2006. PMID: 16820501 Free PMC article.
References
-
- Amati, B., and H. Land. 1994. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation, and death. Curr. Opin. Genet. Dev. 4:102-108. - PubMed
-
- Berteaux-Lecellier, V., M. Picard, C. Thompson-Coffe, D. Zickler, A. Panvier-Adoutte, and J.-M. Simonet. 1995. A nonmammalian homolog of the PAF1 gene (Zellweger Syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell 81:1043-1051. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

