Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae
- PMID: 12455695
- PMCID: PMC126745
- DOI: 10.1128/EC.1.5.774-786.2002
Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae
Abstract
ENG1 (YNR067c), a gene encoding a new endo-1,3-beta-glucanase, was cloned by screening a genomic library with a DNA probe obtained by PCR with synthetic oligonucleotides designed according to conserved regions found between yeast exo-1,3-beta-glucanases (Exglp, Exg2p, and Ssglp). Eng1p shows strong sequence similarity to the product of the Saccharomyces cerevisiae ACF2 gene, involved in actin assembly "in vitro," and to proteins present in other yeast and fungal species. It is also related to plant glucan-binding elicitor proteins, which trigger the onset of a defense response upon fungal infection. Eng1p and Acf2p/Eng2p are glucan-hydrolyzing proteins that specifically act on 1,3-beta linkages, with an endolytic mode of action. Eng1p is an extracellular, heavily glycosylated protein, while Acf2p/Eng2p is an intracellular protein with no carbohydrate linked by N-glycosidic bonds. ENG1 transcription fluctuates periodically during the cell cycle; maximal accumulation occurs during the M/G1 transition and is dependent on the transcription factor Ace2p. Interestingly, eng1 deletion mutants show defects in cell separation, and Eng1p localizes asymmetrically to the daughter side of the septum, suggesting that this protein is involved, together with chitinase, in the dissolution of the mother-daughter septum.
Figures


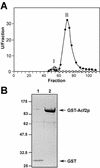


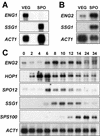

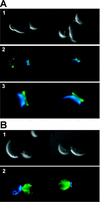
References
-
- Byers, B. 1981. Cytology of the yeast life cycle, p. 59-96. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
-
- Cabib, E., D. H. Roh, M. Schmidt, L. B. Crotti, and A. Varma. 2001. The yeast cell wall and septum as paradigms of cell growth morphogenesis. J. Biol. Chem. 276:19679-19682. - PubMed
-
- Cabib, E., S. J. Silverman, and J. A. Shaw. 1992. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. J. Gen. Microbiol. 138:97-102. - PubMed
-
- Carlson, M., and D. Botstein. 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell 28:145-154. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

