Involvement of RAD9-dependent damage checkpoint control in arrest of cell cycle, induction of cell death, and chromosome instability caused by defects in origin recognition complex in Saccharomyces cerevisiae
- PMID: 12455955
- PMCID: PMC118029
- DOI: 10.1128/EC.1.2.200-212.2002
Involvement of RAD9-dependent damage checkpoint control in arrest of cell cycle, induction of cell death, and chromosome instability caused by defects in origin recognition complex in Saccharomyces cerevisiae
Abstract
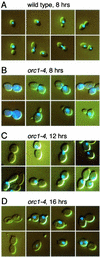
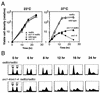
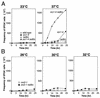
Perturbation of origin firing in chromosome replication is a possible cause of spontaneous chromosome instability in multireplicon organisms. Here, we show that chromosomal abnormalities, including aneuploidy and chromosome rearrangement, were significantly increased in yeast diploid cells with defects in the origin recognition complex. The cell cycle of orc1-4/orc1-4 temperature-sensitive mutant was arrested at the G2/M boundary, after several rounds of cell division at the restrictive temperature. However, prolonged incubation of the mutant cells at 37 degrees C led to abrogation of G2 arrest, and simultaneously the cells started to lose viability. A sharp increase in chromosome instability followed the abrogation of G2 arrest. In orc1-4/orc1-4 rad9delta/rad9delta diploid cells grown at 37 degrees C, G2 arrest and induction of cell death were suppressed, while chromosome instability was synergistically augmented. These findings indicated that DNA lesions caused by a defect in Orc1p function trigger the RAD9-dependent checkpoint control, which ensures genomic integrity either by stopping the cell cycle progress until lesion repair, or by inducing cell death when the lesion is not properly repaired. At semirestrictive temperatures, orc2-1/orc2-1 diploid cells demonstrated G2 arrest and loss of cell viability, both of which require RAD9-dependent checkpoint control. However, chromosome instability was not induced in orc2-1/orc2-1 cells, even in the absence of the checkpoint control. These data suggest that once cells lose the damage checkpoint control, perturbation of origin firing can be tolerated by the cells. Furthermore, although a reduction in origin-firing capacity does not necessarily initiate chromosome instability, the Orc1p possesses a unique function, the loss of which induces instability in the chromosome.
Figures




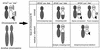
Similar articles
-
Abnormality in initiation program of DNA replication is monitored by the highly repetitive rRNA gene array on chromosome XII in budding yeast.Mol Cell Biol. 2007 Jan;27(2):568-78. doi: 10.1128/MCB.00731-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101800 Free PMC article.
-
Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint.Genetics. 1993 May;134(1):63-80. doi: 10.1093/genetics/134.1.63. Genetics. 1993. PMID: 8514150 Free PMC article.
-
Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair.Genes Dev. 1994 Mar 15;8(6):652-65. doi: 10.1101/gad.8.6.652. Genes Dev. 1994. PMID: 7926756
-
Dual cell cycle checkpoints sensitive to chromosome replication and DNA damage in the budding yeast Saccharomyces cerevisiae.Radiat Res. 1992 Nov;132(2):141-3. Radiat Res. 1992. PMID: 1438694 Review.
-
Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex.DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1009-14. doi: 10.1016/j.dnarep.2004.03.032. DNA Repair (Amst). 2004. PMID: 15279787 Review.
Cited by
-
Abnormality in initiation program of DNA replication is monitored by the highly repetitive rRNA gene array on chromosome XII in budding yeast.Mol Cell Biol. 2007 Jan;27(2):568-78. doi: 10.1128/MCB.00731-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101800 Free PMC article.
-
Pathways and Mechanisms that Prevent Genome Instability in Saccharomyces cerevisiae.Genetics. 2017 Jul;206(3):1187-1225. doi: 10.1534/genetics.112.145805. Genetics. 2017. PMID: 28684602 Free PMC article. Review.
-
Functional connection between the Clb5 cyclin, the protein kinase C pathway and the Swi4 transcription factor in Saccharomyces cerevisiae.Genetics. 2005 Dec;171(4):1485-98. doi: 10.1534/genetics.105.045005. Epub 2005 Aug 22. Genetics. 2005. PMID: 16118191 Free PMC article.
-
Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae.Mol Cell Biol. 2004 Dec;24(23):10208-22. doi: 10.1128/MCB.24.23.10208-10222.2004. Mol Cell Biol. 2004. PMID: 15542831 Free PMC article.
-
Positive and negative roles of homologous recombination in the maintenance of genome stability in Saccharomyces cerevisiae.Genetics. 2003 May;164(1):31-46. doi: 10.1093/genetics/164.1.31. Genetics. 2003. PMID: 12750319 Free PMC article.
References
-
- Bell, S. P., R. Kobayashi, and B. Stillman. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262:1844-1849. - PubMed
-
- Brewer, B. J. 1988. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53:679-686. - PubMed
-
- Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. - PubMed
-
- Bruschi, C. V., J. N. McMillan, M. Coglievina, and M. S. Esposito. 1995. The genomic instability of yeast cdc6-1/cdc6-1 mutants involves chromosome structure and recombination. Mol. Gen. Genet. 249:8-18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

