Role of histone deacetylation in developmentally programmed DNA rearrangements in Tetrahymena thermophila
- PMID: 12455963
- PMCID: PMC118033
- DOI: 10.1128/EC.1.2.293-303.2002
Role of histone deacetylation in developmentally programmed DNA rearrangements in Tetrahymena thermophila
Abstract
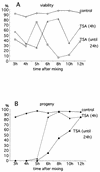
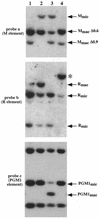
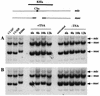
In Tetrahymena, as in other ciliates, development of the somatic macronucleus during conjugation involves extensive and reproducible rearrangements of the germ line genome, including chromosome fragmentation and excision of internal eliminated sequences (IESs). The molecular mechanisms controlling these events are poorly understood. To investigate the role that histone acetylation may play in the regulation of these processes, we treated Tetrahymena cells during conjugation with the histone deacetylase inhibitor trichostatin A (TSA). We show that TSA treatment induces developmental arrests in the early stages of conjugation but does not significantly affect the progression of conjugation once the mitotic divisions of the zygotic nucleus have occurred. Progeny produced from TSA-treated cells were examined for effects on IES excision and chromosome breakage. We found that TSA treatment caused partial inhibition of excision of five out of the six IESs analyzed but did not affect chromosome breakage at four different sites. TSA treatment greatly delayed in some cells and inhibited in most the excision events in the developing macronucleus. It also led to loss of the specialized subnuclear localization of the chromodomain protein Pdd1p that is normally associated with DNA elimination. We propose a model in which underacetylated nucleosomes mark germ line-limited sequences for excision.
Figures


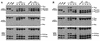

References
-
- Almouzni, G., S. Khochbin, S. Dimitrov, and A. P. Wolffe. 1994. Histone acetylation influences both gene expression and development of Xenopus laevis. Dev. Biol. 165:654-669. - PubMed
-
- Asai, D. J., and J. D. Forney. 2000. Tetrahymena thermophila, vol. 62. Academic Press, San Diego, Calif.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

