A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei
- PMID: 12455980
- PMCID: PMC118050
- DOI: 10.1128/EC.1.1.137-151.2002
A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei
Erratum in
- Eukaryot Cell 2002 Apr;1(2):305
Abstract
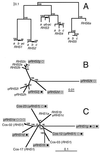
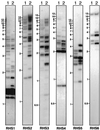
We describe a novel gene family that forms clusters in subtelomeric regions of Trypanosoma brucei chromosomes and partially accounts for the observed clustering of retrotransposons. The ingi and ribosomal inserted mobile element (RIME) non-LTR retrotransposons share 250 bp at both extremities and are the most abundant putatively mobile elements, with about 500 copies per haploid genome. From cDNA clones and subsequently in the T. brucei genomic DNA databases, we identified 52 homologous gene and pseudogene sequences, 16 of which contain a RIME and/or ingi retrotransposon inserted at exactly the same relative position. Here these genes are called the RHS family, for retrotransposon hot spot. Comparison of the protein sequences encoded by RHS genes (21 copies) and pseudogenes (24 copies) revealed a conserved central region containing an ATP/GTP-binding motif and the RIME/ingi insertion site. The RHS proteins share between 13 and 96% identity, and six subfamilies, RHS1 to RHS6, can be defined on the basis of their divergent C-terminal domains. Immunofluorescence and Western blot analyses using RHS subfamily-specific immune sera show that RHS proteins are constitutively expressed and occur mainly in the nucleus. Analysis of Genome Survey Sequence databases indicated that the Trypanosoma brucei diploid genome contains about 280 RHS (pseudo)genes. Among the 52 identified RHS (pseudo)genes, 48 copies are in three RHS clusters located in subtelomeric regions of chromosomes Ia and II and adjacent to the active bloodstream form expression site in T. brucei strain TREU927/4 GUTat10.1. RHS genes comprise the remaining sequence of the size-polymorphic "repetitive region" described for T. brucei chromosome I, and a homologous gene family is present in the Trypanosoma cruzi genome.
Figures


 ), RHS2 (
), RHS2 ( ), RHS3 (▭), RHS4 (
), RHS3 (▭), RHS4 ( ), RHS5 (
), RHS5 ( ), and RHS6 (
), and RHS6 ( ). In the coding sequences, the positions of frame shifts (F), premature stop codons (S), and RIME (○) and/or ingi (•) insertions are indicated. Multiple retroelement insertions are shown by a corresponding number of open and filled circles. Horizontal lines in the middle of ϕRHS1h and ϕRHS2f coding sequences represent a deletion of a part of the RHS coding sequence. The most conserved (box 1) and most divergent (box 2) coding regions between RHS subfamilies are shaded.
). In the coding sequences, the positions of frame shifts (F), premature stop codons (S), and RIME (○) and/or ingi (•) insertions are indicated. Multiple retroelement insertions are shown by a corresponding number of open and filled circles. Horizontal lines in the middle of ϕRHS1h and ϕRHS2f coding sequences represent a deletion of a part of the RHS coding sequence. The most conserved (box 1) and most divergent (box 2) coding regions between RHS subfamilies are shaded.
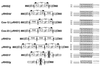



References
-
- Affolter, M., L. Rindisbacher, and R. Braun. 1989. The tubulin gene cluster of Trypanosoma brucei starts with an intact beta-gene and ends with a truncated beta-gene interrupted by a retrotransposon-like sequence. Gene 80:177-183. - PubMed
-
- Barry, J. D., S. V. Graham, M. Fotheringham, V. S. Graham, K. Kobryn, and B. Wymer. 1998. VSG gene control and infectivity strategy of metacyclic stage Trypanosoma brucei. Mol. Biochem. Parasitol. 91:93-105. - PubMed
-
- Barry, J. D., and R. McCulloch. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49:1-70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

