A G-protein beta subunit required for sexual and vegetative development and maintenance of normal G alpha protein levels in Neurospora crassa
- PMID: 12455986
- PMCID: PMC118013
- DOI: 10.1128/EC.1.3.378-390.2002
A G-protein beta subunit required for sexual and vegetative development and maintenance of normal G alpha protein levels in Neurospora crassa
Abstract
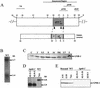
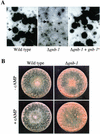
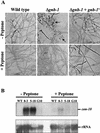
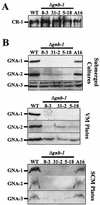
The genome of the filamentous fungus Neurospora crassa contains a single gene encoding a heterotrimeric G-protein beta subunit, gnb-1. The predicted GNB-1 protein sequence is most identical to G beta proteins from the filamentous fungi Cryphonectria parasitica and Aspergillus nidulans. N. crassa GNB-1 is also 65% identical to the human GNB-1 protein but only 38 and 45% identical to G beta proteins from budding and fission yeasts. Previous studies in animal and fungal systems have elucidated phenotypes of G beta null mutants, but little is known about the effects of G beta loss on G alpha levels. In this study, we analyzed a gnb-1 deletion mutant for cellular phenotypes and levels of the three G alpha proteins. Delta gnb-1 strains are female-sterile, with production of aberrant fertilized reproductive structures. Delta gnb-1 strains conidiate more profusely and have altered mass on solid medium. Loss of gnb-1 leads to inappropriate conidiation and expression of a conidiation-specific gene during growth in submerged culture. Intracellular cyclic AMP levels are reduced by 60% in vegetative plate cultures of delta gnb-1 mutants. Loss of gnb-1 leads to lower levels of the three G alpha proteins under a variety of conditions. Analysis of transcript levels for the gna-1 and gna-2 G alpha genes in submerged cultures indicates that regulation of G alpha protein levels by gnb-1 is posttranscriptional. The results suggest that GNB-1 directly regulates apical extension rate and mass accumulation. In contrast, many other delta gnb-1 phenotypes, including female sterility and defective conidiation, can be explained by altered levels of the three N. crassa G alpha proteins.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

