Differential modulation of cellular signaling pathways by mild and severe hypovirus strains
- PMID: 12455988
- PMCID: PMC118008
- DOI: 10.1128/EC.1.3.401-413.2002
Differential modulation of cellular signaling pathways by mild and severe hypovirus strains
Abstract
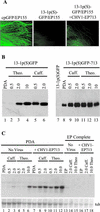
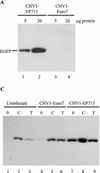
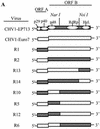
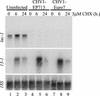
Hypoviruses persistently alter multiple phenotypic traits, stably modify gene expression, and attenuate virulence (hypovirulence) of their pathogenic fungal host, the chestnut blight fungus Cryphonectria parasitica. The pleiotropic nature of these changes is consistent with hypovirus-mediated perturbation of one or more cellular signal transduction pathways. We now report that two hypoviruses that differ in the severity of symptom expression differentially perturb specific cellular signaling pathways. The C. parasitica 13-1 gene, originally identified as a hypovirus-inducible and cyclic AMP (cAMP)-regulated gene, was used to design a promoter-GFP reporter construct with which to monitor perturbation of cAMP-mediated signaling. Virus-mediated modulation of calcium/calmodulin/inositol trisphosphate-dependent signaling was monitored by measuring transcript accumulation from the C. parasitica laccase gene, lac-1. Infection by the severe hypovirus strain CHV1-EP713 caused a substantial induction of 13-1 promoter activity and a reduction of total extracellular laccase enzymatic activity (LAC-1 and LAC-3). In contrast, 13-1 promoter activity and total laccase activity were only marginally altered upon infection with the mild hypovirus strain CHV1-Euro7. However, examination of lac-1-specific transcript accumulation under previously defined culture conditions revealed that both CHV1-EP713 and CHV1-Euro7 perturbed calcium/calmodulin/inositol trisphosphate-dependent signaling. CHV1-EP713/CHV1-Euro7 chimeric viruses were used to map viral determinants responsible for modulation of cAMP-dependent signaling to domains within the central portion of the second open reading frame.
Figures









References
-
- Anagnostakis, S. L. 1982. Biological control of chestnut blight. Science 215:466-471. - PubMed
-
- Anagnostakis, S. L. 1984. Nuclear gene mutations in Endothia (Cryphonectria) parasitica that affect morphology and virulence. Phytopathology 74:561-565.
-
- Anagnostakis, S. L., B. Chen, L. M. Geletka, and D. L. Nuss. 1998. Hypovirus transmission to ascospore progeny by field-released transgenic hypovirulent strains of Cryphonectria parasitica. Phytopathology 88:598-604. - PubMed
-
- Anagnostakis, S. L., and P. R. Day. 1979. Hypovirulence conversion in Endothia parasitica. Phytopathology 69:1226-1229.
-
- Bavendamm, W. 1928. Ueber das Vorkommen und den Nachweis von Oxydasen bei holzzerstörenden Pilzen. Z. Pflanzenschutz 38:257-276.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

