GTPase-activating proteins for Cdc42
- PMID: 12455995
- PMCID: PMC118018
- DOI: 10.1128/EC.1.3.469-480.2002
GTPase-activating proteins for Cdc42
Abstract
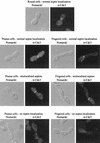
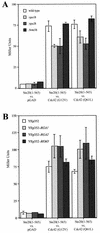
The Rho-type GTPase, Cdc42, has been implicated in a variety of functions in the yeast life cycle, including septin organization for cytokinesis, pheromone response, and haploid invasive growth. A group of proteins called GTPase-activating proteins (GAPs) catalyze the hydrolysis of GTP to GDP, thereby inactivating Cdc42. At the time this study began, there was one known GAP, Bem3, and one putative GAP, Rga1, for Cdc42. We identified another putative GAP for Cdc42 and named it Rga2 (Rho GTPase-activating protein 2). We confirmed by genetic and biochemical criteria that Rga1, Rga2, and Bem3 act as GAPs for Cdc42. A detailed characterization of Rga1, Rga2, and Bem3 suggested that they regulate different subsets of Cdc42 function. In particular, deletion of the individual GAPs conferred different phenotypes. For example, deletion of RGA1, but not RGA2 or BEM3, caused hyperinvasive growth. Furthermore, overproduction or loss of Rga1 and Rga2, but not Bem3, affected the two-hybrid interaction of Cdc42 with Ste20, a p21-activated kinase (PAK) kinase required for haploid invasive growth. These results suggest Rga1, and possibly Rga2, facilitate the interaction of Cdc42 with Ste20 to mediate signaling in the haploid invasive growth pathway. Deletion of BEM3 resulted in cells with severe morphological defects not observed in rga1delta or rga2delta strains. These data suggest that Bem3 and, to a lesser extent, Rga1 and Rga2 facilitate the role of Cdc42 in septin organization. Thus, it appears that the GAPs play a role in modulating specific aspects of Cdc42 function. Alternatively, the different phenotypes could reflect quantitative rather than qualitative differences in GAP activity in the mutant strains.
Figures


Similar articles
-
Roles of an N-terminal coiled-coil-containing domain in the localization and function of Bem3, a Rho GTPase-activating protein in budding yeast.Fungal Genet Biol. 2017 Feb;99:40-51. doi: 10.1016/j.fgb.2016.12.010. Epub 2017 Jan 4. Fungal Genet Biol. 2017. PMID: 28064039
-
Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain.J Biol Chem. 1993 Nov 25;268(33):24629-34. J Biol Chem. 1993. PMID: 8227021
-
Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae.Genes Dev. 1995 Dec 1;9(23):2949-63. doi: 10.1101/gad.9.23.2949. Genes Dev. 1995. PMID: 7498791
-
Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways.Eur J Biochem. 1996 Dec 1;242(2):171-85. doi: 10.1111/j.1432-1033.1996.0171r.x. Eur J Biochem. 1996. PMID: 8973630 Review.
-
Complexity and self-organization in the evolution of cell polarization.J Cell Sci. 2023 Jan 15;136(2):jcs259639. doi: 10.1242/jcs.259639. Epub 2023 Jan 24. J Cell Sci. 2023. PMID: 36691920 Review.
Cited by
-
Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p.Mol Biol Cell. 2007 Jun;18(6):2090-101. doi: 10.1091/mbc.e06-09-0883. Epub 2007 Mar 21. Mol Biol Cell. 2007. PMID: 17377067 Free PMC article.
-
CDC42 Regulatory Patterns Related To Inflammatory Bowel Disease and Hyperglycemia.J Bioinform Syst Biol. 2025;8(1):17-28. Epub 2025 Feb 20. J Bioinform Syst Biol. 2025. PMID: 40183002 Free PMC article.
-
A protein complex containing Epo1p anchors the cortical endoplasmic reticulum to the yeast bud tip.J Cell Biol. 2015 Jan 5;208(1):71-87. doi: 10.1083/jcb.201407126. Epub 2014 Dec 29. J Cell Biol. 2015. PMID: 25547157 Free PMC article.
-
Morphogenesis and the cell cycle.Genetics. 2012 Jan;190(1):51-77. doi: 10.1534/genetics.111.128314. Genetics. 2012. PMID: 22219508 Free PMC article. Review.
-
The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast.Mol Biol Cell. 2003 Oct;14(10):4051-66. doi: 10.1091/mbc.e03-04-0247. Epub 2003 Jul 25. Mol Biol Cell. 2003. PMID: 14517318 Free PMC article.
References
-
- Barthe, C., G. de Bettignies, O. Louvet, M. F. Peypouquet, C. Morel, F. Doignon, and M. Crouzet. 1998. First characterization of the gene RGD1 in the yeast Saccharomyces cerevisiae. C. R. Acad. Sci. Ser. III 321:453-462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

