Isolation and characterization of YlBEM1, a gene required for cell polarization and differentiation in the dimorphic yeast Yarrowia lipolytica
- PMID: 12456001
- PMCID: PMC118001
- DOI: 10.1128/EC.1.4.526-537.2002
Isolation and characterization of YlBEM1, a gene required for cell polarization and differentiation in the dimorphic yeast Yarrowia lipolytica
Abstract
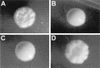
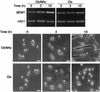
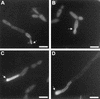
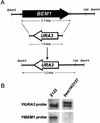
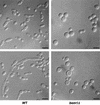
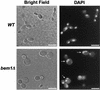
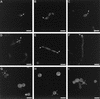
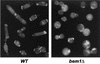
The ability to switch between a unicellular yeast form and different filamentous forms (fungal dimorphism) is an important attribute of most pathogenic fungi. Dimorphism involves a series of events that ultimately result in dramatic changes in the polarity of cell growth in response to environmental factors. We have isolated and characterized YlBEM1, a gene encoding a protein of 639 amino acids that is essential for the yeast-to-hypha transition in the yeast Yarrowia lipolytica and whose transcription is significantly increased during this event. Cells with deletions of YlBEM1 are viable but show substantial alterations in morphology, disorganization of the actin cytoskeleton, delocalization of cortical actin and chitin deposition, multinucleation, and loss of mating ability, thus pointing to a major role for YlBEM1 in the regulation of cell polarity and morphogenesis in this fungus. This role is further supported by the localization of YlBemlp, which, like cortical actin, appears to be particularly abundant at sites of growth of yeast, hyphal, and pseudohyphal cells. In addition, the potential involvement of YlBem1p in septum formation and/or cytokinesis is suggested by the concentration of a green fluorescent protein-tagged version of this protein at the mother-bud neck during the last stages of cell division. Interestingly, overexpression of MHY1, YlRAC1, or YlSEC31, three genes involved in filamentous growth of Y. lipolytica, induced hyphal growth of bem1 null mutant cells.
Figures



References
-
- Adams, A. E. M., and J. R. Pringle. 1991. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 194:729-731. - PubMed
-
- Adisa, A., F. R. Albano, J. Reeder, M. Foley, and L. Tilley. 2001. Evidence for a role for a Plasmodium falciparum homologue of Sec31p in the export of proteins to the surface of malaria parasite-infected erythrocytes. J. Cell Sci. 114:3377-3386. - PubMed
-
- Ausubel, F. J., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Greene Publishing Associates, New York, N.Y.
-
- Banuett, F. 1995. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genet. 29:179-208. - PubMed
-
- Barth, G., and H. Weber. 1986. Improvement of sporulation in the yeast Yarrowia lipolytica. Antonie Leewenhoek J. Microbiol. Serol. 51:167-177. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

