New class of cargo protein in Tetrahymena thermophila dense core secretory granules
- PMID: 12456006
- PMCID: PMC117993
- DOI: 10.1128/EC.1.4.583-593.2002
New class of cargo protein in Tetrahymena thermophila dense core secretory granules
Abstract
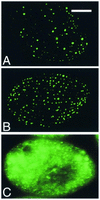
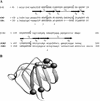
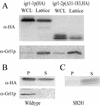
Regulated exocytosis of dense core secretory granules releases biologically active proteins in a stimulus-dependent fashion. The packaging of the cargo within newly forming granules involves a transition: soluble polypeptides condense to form water-insoluble aggregates that constitute the granule cores. Following exocytosis, the cores generally disassemble to diffuse in the cell environment. The ciliates Tetrahymena thermophila and Paramecium tetraurelia have been advanced as genetically manipulatable systems for studying exocytosis via dense core granules. However, all of the known granule proteins in these organisms condense to form the architectural units of lattices that are insoluble both before and after exocytosis. Using an approach designed to detect new granule proteins, we have now identified Igr1p (induced during granule regeneration). By structural criteria, it is unrelated to the previously characterized lattice-forming proteins. It is distinct in that it is capable of dissociating from the insoluble lattice following secretion and therefore represents the first diffusible protein identified in ciliate granules.
Figures




References
-
- Chan, C. W., Y. Saimi, and C. Kung. 1999. A new multigene family encoding calcium-dependent calmodulin-binding membrane proteins of Paramecium tetraurelia. Gene 231:21-32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

