The fission yeast ubiquitin-conjugating enzymes UbcP3, Ubc15, and Rhp6 affect transcriptional silencing of the mating-type region
- PMID: 12456009
- PMCID: PMC118003
- DOI: 10.1128/EC.1.4.613-625.2002
The fission yeast ubiquitin-conjugating enzymes UbcP3, Ubc15, and Rhp6 affect transcriptional silencing of the mating-type region
Abstract
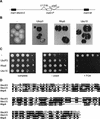
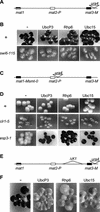
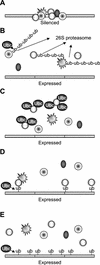
Genes transcribed by RNA polymerase II are silenced when introduced near the mat2 or mat3 mating-type loci of the fission yeast Schizosaccharomyces pombe. Silencing is mediated by a number of gene products and cis-acting elements. We report here the finding of novel trans-acting factors identified in a screen for high-copy-number disruptors of silencing. Expression of cDNAs encoding the putative E2 ubiquitin-conjugating enzymes UbcP3, Ubc15 (ubiquitin-conjugating enzyme), or Rhp6 (Rad homolog pombe) from the strong nmt1 promoter derepressed the silent mating-type loci mat2 and mat3 and reporter genes inserted nearby. Deletion of rhp6 slightly derepressed an ade6 reporter gene placed in the mating-type region, whereas disruption of ubcP3 or ubc15 had no obvious effect on silencing. Rhp18 is the S. pombe homolog of Saccharomyces cerevisiae Rad18p, a DNA-binding protein that physically interacts with Rad6p. Rhp18 was not required for the derepression observed when UbcP3, Ubc15, or Rhp6 was overproduced. Overexpressing Rhp6 active-site mutants showed that the ubiquitin-conjugating activity of Rhp6 is essential for disruption of silencing. However, high dosage of UbcP3, Ubc15, or Rhp6 was not suppressed by a mutation in the 26S proteasome, suggesting that loss of silencing is not due to an increased degradation of silencing factors but rather to the posttranslational modification of proteins by ubiquitination. We discuss the implications of these results for the possible modes of action of UbcP3, Ubc15, and Rhp6.
Figures






Similar articles
-
Two ubiquitin-conjugating enzymes, Rhp6 and UbcX, regulate heterochromatin silencing in Schizosaccharomyces pombe.Mol Cell Biol. 2002 Dec;22(23):8366-74. doi: 10.1128/MCB.22.23.8366-8374.2002. Mol Cell Biol. 2002. PMID: 12417737 Free PMC article.
-
Identification of Uhp1, a ubiquitinated histone-like protein, as a target/mediator of Rhp6 in mating-type silencing in fission yeast.J Biol Chem. 2003 Mar 14;278(11):9185-94. doi: 10.1074/jbc.M212732200. Epub 2003 Jan 2. J Biol Chem. 2003. PMID: 12511578
-
A novel function of the DNA repair gene rhp6 in mating-type silencing by chromatin remodeling in fission yeast.Mol Cell Biol. 1998 Sep;18(9):5511-22. doi: 10.1128/MCB.18.9.5511. Mol Cell Biol. 1998. PMID: 9710635 Free PMC article.
-
Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe.Nucleic Acids Res. 2002 Apr 1;30(7):1465-82. doi: 10.1093/nar/30.7.1465. Nucleic Acids Res. 2002. PMID: 11917007 Free PMC article. Review.
-
UBA domain containing proteins in fission yeast.Int J Biochem Cell Biol. 2003 May;35(5):629-36. doi: 10.1016/s1357-2725(02)00393-x. Int J Biochem Cell Biol. 2003. PMID: 12672455 Review.
Cited by
-
Interaction of APC/C-E3 ligase with Swi6/HP1 and Clr4/Suv39 in heterochromatin assembly in fission yeast.J Biol Chem. 2009 Mar 13;284(11):7165-76. doi: 10.1074/jbc.M806461200. Epub 2008 Dec 30. J Biol Chem. 2009. PMID: 19117951 Free PMC article.
-
Concerted action of the ubiquitin-fusion degradation protein 1 (Ufd1) and Sumo-targeted ubiquitin ligases (STUbLs) in the DNA-damage response.PLoS One. 2013 Nov 12;8(11):e80442. doi: 10.1371/journal.pone.0080442. eCollection 2013. PLoS One. 2013. PMID: 24265825 Free PMC article.
-
The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe.Genetics. 2005 Dec;171(4):1583-95. doi: 10.1534/genetics.105.048298. Epub 2005 Sep 12. Genetics. 2005. PMID: 16157682 Free PMC article.
-
A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation.Genes Dev. 2005 Jul 15;19(14):1705-14. doi: 10.1101/gad.1328005. Genes Dev. 2005. PMID: 16024659 Free PMC article.
-
Replication stress affects the fidelity of nucleosome-mediated epigenetic inheritance.PLoS Genet. 2017 Jul 27;13(7):e1006900. doi: 10.1371/journal.pgen.1006900. eCollection 2017 Jul. PLoS Genet. 2017. PMID: 28749973 Free PMC article.
References
-
- Allshire, R. C., J. P. Javerzat, N. J. Redhead, and G. Cranston. 1994. Position effect variegation at fission yeast centromeres. Cell 76:157-169. - PubMed
-
- Allshire, R. C. 1996. Transcriptional silencing in the fission yeast: a manifestation of higher order chromosome structure and function, p. 443-466. In V. E. A. Russo, R. A. Martienssen, and A. D. Riggs (ed.), Epigenetic mechanisms of gene regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
-
- Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

