A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B
- PMID: 12456659
- PMCID: PMC136951
- DOI: 10.1093/emboj/cdf647
A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B
Abstract
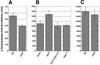
Import of core histones into the nucleus is a prerequisite for their deposition onto DNA and the assembly of chromatin. Here we demonstrate that nucleosome assembly protein 1 (Nap1p), a protein previously implicated in the deposition of histones H2A and H2B, is also involved in the transport of these two histones. We demonstrate that Nap1p can bind directly to Kap114p, the primary karyopherin/importin responsible for the nuclear import of H2A and H2B. Nap1p also serves as a bridge between Kap114p and the histone nuclear localization sequence (NLS). Nap1p acts cooperatively to increase the affinity of Kap114p for these NLSs. Nuclear accumulation of histone NLS-green fluorescent protein (GFP) reporters was decreased in deltanap1 cells. Furthermore, we demonstrate that Nap1p promotes the association of the H2A and H2B NLSs specifically with the karyopherin Kap114p. Localization studies demonstrate that Nap1p is a nucleocytoplasmic shuttling protein, and genetic experiments suggest that its shuttling is important for maintaining chromatin structure in vivo. We propose a model in which Nap1p links the nuclear transport of H2A and H2B to chromatin assembly.
Figures





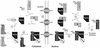
Similar articles
-
Nuclear import of histone H2A and H2B is mediated by a network of karyopherins.J Cell Biol. 2001 Apr 16;153(2):251-62. doi: 10.1083/jcb.153.2.251. J Cell Biol. 2001. PMID: 11309407 Free PMC article.
-
Modulation of histone deposition by the karyopherin kap114.Mol Cell Biol. 2005 Mar;25(5):1764-78. doi: 10.1128/MCB.25.5.1764-1778.2005. Mol Cell Biol. 2005. PMID: 15713633 Free PMC article.
-
Nuclear import of TFIIB is mediated by Kap114p, a karyopherin with multiple cargo-binding domains.Mol Biol Cell. 2005 Jul;16(7):3200-10. doi: 10.1091/mbc.e04-11-0990. Epub 2005 May 11. Mol Biol Cell. 2005. PMID: 15888545 Free PMC article.
-
Nuclear import of histones.Biochem Soc Trans. 2020 Dec 18;48(6):2753-2767. doi: 10.1042/BST20200572. Biochem Soc Trans. 2020. PMID: 33300986 Free PMC article. Review.
-
Chromatin assembly: biochemical identities and genetic redundancy.Curr Opin Genet Dev. 1999 Apr;9(2):185-90. doi: 10.1016/S0959-437X(99)80028-8. Curr Opin Genet Dev. 1999. PMID: 10322140 Review.
Cited by
-
Molecular Size Analysis of Recombinant Importin-histone Complexes Using Analytical Ultracentrifugation.Bio Protoc. 2020 May 20;10(10):e3625. doi: 10.21769/BioProtoc.3625. eCollection 2020 May 20. Bio Protoc. 2020. PMID: 33659298 Free PMC article.
-
Structural convergence endows nuclear transport receptor Kap114p with a transcriptional repressor function toward TATA-binding protein.Nat Commun. 2023 Sep 8;14(1):5518. doi: 10.1038/s41467-023-41206-9. Nat Commun. 2023. PMID: 37684250 Free PMC article.
-
Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions.Traffic. 2010 Feb;11(2):185-97. doi: 10.1111/j.1600-0854.2009.01010.x. Epub 2009 Nov 19. Traffic. 2010. PMID: 19929865 Free PMC article.
-
Histone chaperones in nucleosome assembly and human disease.Nat Struct Mol Biol. 2013 Jan;20(1):14-22. doi: 10.1038/nsmb.2461. Nat Struct Mol Biol. 2013. PMID: 23288364 Free PMC article. Review.
-
Histone chaperones Nap1 and Vps75 regulate histone acetylation during transcription elongation.Mol Cell Biol. 2013 Apr;33(8):1645-56. doi: 10.1128/MCB.01121-12. Epub 2013 Feb 11. Mol Cell Biol. 2013. PMID: 23401858 Free PMC article.
References
-
- Adams C.R. and Kamakaka,R.T. (1999) Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev., 9, 185–190. - PubMed
-
- Aitchison J.D., Blobel,G. and Rout,M.P. (1996) Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science, 274, 624–627. - PubMed
-
- Bagley S., Goldberg,M.W., Cronshaw,J.M., Rutherford,S. and Allen,T.D. (2000) The nuclear pore complex. J. Cell Sci., 113, 3885–3886. - PubMed
-
- Chang L., Loranger,S.S., Mizzen,C., Ernst,S.G., Allis,C.D. and Annunziato,A.T. (1997) Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry, 36, 469–480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

