Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent
- PMID: 12456788
- PMCID: PMC134655
- DOI: 10.1128/MMBR.66.4.702-738.2002
Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent
Abstract
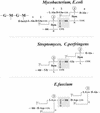
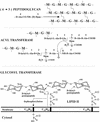
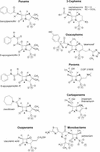
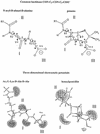
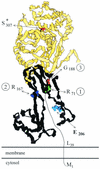
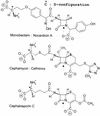
The bacterial acyltransferases of the SxxK superfamily vary enormously in sequence and function, with conservation of particular amino acid groups and all-alpha and alpha/beta folds. They occur as independent entities (free-standing polypeptides) and as modules linked to other polypeptides (protein fusions). They can be classified into three groups. The group I SxxK D,D-acyltransferases are ubiquitous in the bacterial world. They invariably bear the motifs SxxK, SxN(D), and KT(S)G. Anchored in the plasma membrane with the bulk of the polypeptide chain exposed on the outer face of it, they are implicated in the synthesis of wall peptidoglycans of the most frequently encountered (4-->3) type. They are inactivated by penicillin and other beta-lactam antibiotics acting as suicide carbonyl donors in the form of penicillin-binding proteins (PBPs). They are components of a morphogenetic apparatus which, as a whole, controls multiple parameters such as shape and size and allows the bacterial cells to enlarge and duplicate their particular pattern. Class A PBP fusions comprise a glycosyltransferase module fused to an SxxK acyltransferase of class A. Class B PBP fusions comprise a linker, i.e., protein recognition, module fused to an SxxK acyltransferase of class B. They ensure the remodeling of the (4-->3) peptidoglycans in a cell cycle-dependent manner. The free-standing PBPs hydrolyze D,D peptide bonds. The group II SxxK acyltransferases frequently have a partially modified bar code, but the SxxK motif is invariant. They react with penicillin in various ways and illustrate the great plasticity of the catalytic centers. The secreted free-standing PBPs, the serine beta-lactamases, and the penicillin sensors of several penicillin sensory transducers help the D,D-acyltransferases of group I escape penicillin action. The group III SxxK acyltransferases are indistinguishable from the PBP fusion proteins of group I in motifs and membrane topology, but they resist penicillin. They are referred to as Pen(r) protein fusions. Plausible hypotheses are put forward on the roles that the Pen(r) protein fusions, acting as L,D-acyltransferases, may play in the (3-->3) peptidoglycan-synthesizing molecular machines. Shifting the wall peptidoglycan from the (4-->3) type to the (3-->3) type could help Mycobacterium tuberculosis and Mycobacterium leprae survive by making them penicillin resistant.
Figures

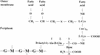


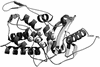







References
-
- Adam, M., C. Fraipont, N. Rhazi, M. Nguyen-Distèche, B. Lakaye, J. M. Frère, B. Devreese, J. Van Beeumen, Y. van Heijenoort, J. van Heijenoort, and J. M. Ghuysen. 1997. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia coli catalyzes peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from lipid II intermediate. J. Bacteriol. 179:6005-6009. - PMC - PubMed
-
- Addinall, S. G., and J. Lutkenhaus. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22:231-237. - PubMed
-
- Altschul, S. F., and W. Gish. 1996. Local alignment statistics. Methods Enzymol. 266:460-480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

