Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency
- PMID: 12456880
- PMCID: PMC138598
- DOI: 10.1073/pnas.252630899
Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency
Abstract
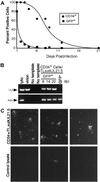
Human cytomegalovirus (HCMV) resides latently in hematopoietic cells of the bone marrow. Although viral genomes can be found in CD14+ monocytes and CD34+ progenitor cells, the primary reservoir for latent cytomegalovirus is unknown. We analyzed human hematopoietic subpopulations infected in vitro with a recombinant virus that expresses a green fluorescent protein marker gene. Although many hematopoietic cell subsets were infected in vitro, CD14+ monocytes and various CD34+ subpopulations were infected with the greatest efficiency. We have developed an in vitro system in which to study HCMV infection and latency in CD34+ cells cultured with irradiated stromal cells. Marker gene expression was substantially reduced by 4 days postinfection, and infectious virus was not made during the culture period. However, viral DNA sequences were maintained in infected CD34+ cells for >20 days in culture, and, importantly, virus replication could be reactivated by coculture with human fibroblasts. Using an HCMV gene array, we examined HCMV gene expression in CD34+ cells. The pattern of viral gene expression was distinct from that observed during productive or nonproductive infections. Some of these expressed viral genes may function in latency and are targets for further analysis. Altered gene expression in hematopoietic progenitors may be indicative of the nature and outcome of HCMV infection.
Figures

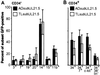


References
-
- Britt W. J. & Alford, C. A. (1996) in Fields Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott–Raven, Philadelphia), Vol. 2, pp. 2493–2524.
-
- Sinzger C., Grefte, A., Plachter, B., Gouw, A. S., The, T. H. & Jahn, G. (1995) J. Gen. Virol. 76, 741-750. - PubMed
-
- Schrier R. D., Nelson, J. A. & Oldstone, M. B. (1985) Science 230, 1048-1051. - PubMed
-
- Boeckh M., Hoy, C. & Torok-Storb, B. (1998) Clin. Infect. Dis. 26, 209-210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

