Extension and integration of the gene ontology (GO): combining GO vocabularies with external vocabularies
- PMID: 12466303
- PMCID: PMC187579
- DOI: 10.1101/gr.580102
Extension and integration of the gene ontology (GO): combining GO vocabularies with external vocabularies
Abstract
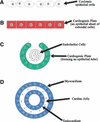
Structured vocabulary development enhances the management of information in biological databases. As information grows, handling the complexity of vocabularies becomes difficult. Defined methods are needed to manipulate, expand and integrate complex vocabularies. The Gene Ontology (GO) project provides the scientific community with a set of structured vocabularies to describe domains of molecular biology. The vocabularies are used for annotation of gene products and for computational annotation of sequence data sets. The vocabularies focus on three concepts universal to living systems, biological process, molecular function and cellular component. As the vocabularies expand to incorporate terms needed by diverse annotation communities, species-specific terms become problematic. In particular, the use of species-specific anatomical concepts remains unresolved. We present a method for expansion of GO into areas outside of the three original universal concept domains. We combine concepts from two orthogonal vocabularies to generate a larger, more specific vocabulary. The example of mammalian heart development is presented because it addresses two issues that challenge GO; inclusion of organism-specific anatomical terms, and proliferation of terms and relationships. The combination of concepts from orthogonal vocabularies provides a robust representation of relevant terms and an opportunity for evaluation of hypothetical concepts.
Figures







References
-
- Abdelwahid E, Pelliniemi LJ, Niinikoski H, Simell O, Tuominen J, Rahkonen O, Jokinen E. Apoptosis in the pattern formation of the ventricular wall during mouse heart organogenesis. Anat Rec. 1999;256:208–217. - PubMed
-
- Aho AV, Hopcroft JE, Ullman JD. Data structures and algorithms. Reading, MA: Addison-Wesley; 1983. Directed graphs; pp. 219–221.
-
- Ashburner M, Lewis S. In silico biology. Novartis Symposium. 2002. On ontologies for biologists: The Gene Ontology—Uncoupling the web. (in press). - PubMed
-
- Bard JBL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock R, Davidson DR. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech Dev. 1998;74:111–120. - PubMed
-
- Berman JJ, Moore GW. SNOMED-encoded surgical pathology databases: A tool for epidemiologic investigation. Mod Pathol. 1996;9:944–950. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
