Skip interacts with the retinoblastoma tumor suppressor and inhibits its transcriptional repression activity
- PMID: 12466551
- PMCID: PMC137971
- DOI: 10.1093/nar/gkf658
Skip interacts with the retinoblastoma tumor suppressor and inhibits its transcriptional repression activity
Abstract
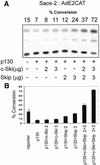
Ski interacting protein (Skip) plays an important role in the transforming activity of both v-Ski and EBNA2 (Epstein-Barr virus encoded latency protein) and is involved in EBNA2 and NotchIC activation of CBF1-repressed promoters. We have previously shown that Skip acts as a transcriptional co-activator on a number of cellular and viral promoters. Here, we report that Skip also interacts with pRb and, in cooperation with Ski, can overcome pRb-induced transcriptional repression. We show a strong and direct interaction between pRb and Skip, and we map the site of interaction to amino acid residues 171-353 of the evolutionarily conserved SNW domain of Skip. Furthermore, the combination of Skip and Ski can successfully overcome the G1 arrest and flat cell phenotype induced by pRb. Taken together, these studies suggest that one potential function of the Skip-Ski complex is to overcome the growth-suppressive activities of pRb.
Figures









References
-
- Folk P., Puta,F., Krpejsova,L., Blahuskova,A., Markos,A., Rabino,M. and Dottin,R.P. (1996) The homolog of chromatin binding protein Bx42 identified in Dictyostelium. Gene, 181, 229–231. - PubMed
-
- Dahl R., Wani,B. and Hayman,M.J. (1998) The Ski oncoprotein interacts with Skip, the human homologue of Drosophila Bx42. Oncogene, 16, 1579–1586. - PubMed
-
- Colmenares C. and Stavnezer,E. (1989) The ski oncogene induces muscle differentiation in quail embryo cells. Cell, 59, 293–303. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

