PSKH1, a novel splice factor compartment-associated serine kinase
- PMID: 12466556
- PMCID: PMC137962
- DOI: 10.1093/nar/gkf648
PSKH1, a novel splice factor compartment-associated serine kinase
Abstract
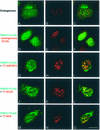
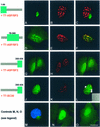
Small nuclear ribonucleoprotein particles (snRNPs) and non-snRNP splicing factors containing a serine/arginine-rich domain (SR proteins) concentrate in splicing factor compartments (SFCs) within the nucleus of interphase cells. Nuclear SFCs are considered mainly as storage sites for splicing factors, supplying splicing factors to active genes. The mechanisms controlling the interaction of the various spliceosome constituents, and the dynamic nature of the SFCs, are still poorly understood. We show here that endogenous PSKH1, a previously cloned kinase, is located in SFCs. Migration of PSKH1-FLAG into SFCs is enhanced during co-expression of T7-tagged ASF/SF2 as well as other members of the SR protein family, but not by two other non-SR nuclear proteins serving as controls. Similar to the SR protein kinase family, overexpression of PSKH1 led to reorganization of co-expressed T7-SC35 and T7-ASF/SF2 into a more diffuse nuclear pattern. This redistribution was not dependent on PSKH1 kinase activity. Different from the SR protein kinases, the SFC-associating features of PSKH1 were located within its catalytic kinase domain and within its C-terminus. Although no direct interaction was observed between PSKH1 and any of the SR proteins tested in pull-down or yeast two-hybrid assays, forced expression of PSKH1-FLAG was shown to stimulate distal splicing of an E1A minigene in HeLa cells. Moreover, a GST-ASF/SF2 fusion was not phosphorylated by PSKH1, suggesting an indirect mechanism of action on SR proteins. Our data suggest a mutual relationship between PSKH1 and SR proteins, as they are able to target PSKH1 into SFCs, while forced PSKH1 expression modulates nuclear dynamics and the function of co-expressed splicing factors.
Figures



References
-
- Jiang Z.H. and Wu,J.Y. (1999) Alternative splicing and programmed cell death. Proc. Soc. Exp. Biol. Med., 220, 64–72. - PubMed
-
- Skalsky Y.M., Ajuh,P.M., Parker,C., Lamond,A.I., Goodwin,G. and Cooper,C.S. (2001) PRCC, the commonest TFE3 fusion partner in papillary renal carcinoma is associated with pre-mRNA splicing factors. Oncogene, 11, 178–187. - PubMed
-
- Rio D.C. (1993) Splicing of pre-mRNA: mechanism, regulation and role in development. Curr. Opin. Genet. Dev., 3, 574–584. - PubMed
-
- Berget S.M. (1995) Exon recognition in vertebrate splicing. J. Biol. Chem., 270, 2411–2414. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

