Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH
- PMID: 12473685
- PMCID: PMC2173389
- DOI: 10.1083/jcb.200207115
Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH
Abstract
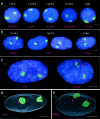
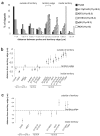
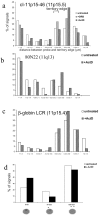
Genes can be transcribed from within chromosome territories; however, the major histocompatibilty complex locus has been reported extending away from chromosome territories, and the incidence of this correlates with transcription from the region. A similar result has been seen for the epidermal differentiation complex region of chromosome 1. These data suggested that chromatin decondensation away from the surface of chromosome territories may result from, and/or may facilitate, transcription of densely packed genes subject to coordinate regulation.To investigate whether localization outside of the visible confines of chromosome territories can also occur for regions that are not coordinately regulated, we have examined the spatial organization of human 11p15.5 and the syntenic region on mouse chromosome 7. This region is gene rich but its genes are not coordinately expressed, rather overall high levels of transcription occur in several cell types. We found that chromatin from 11p15.5 frequently extends away from the chromosome 11 territory. Localization outside of territories was also detected for other regions of high gene density and high levels of transcription. This is shown to be partly dependent on ongoing transcription. We suggest that local gene density and transcription, rather than the activity of individual genes, influences the organization of chromosomes in the nucleus.
Figures



References
-
- Alders, M., M. Hodges, A.K. Hadjantonakis, J. Postmus, I. van Wijk, J. Bliek, M. de Meulemeester, A. Westerveld, F. Guillemot, C. Oudejans, P. Little, M. Mannens. 1997. The human Achaete-Scute homologue 2 (ASCL2,HASH2) maps to chromosome 11p15.5, close to IGF2 and is expressed in extravillus trophoblasts. Hum. Mol. Genet. 6:859–867. - PubMed
-
- Bepler, G., K.C. O'Briant, Y.C. Kim, G. Schreiber, and D.M. Pitterle. 1999. A 1.4-Mb high-resolution physical map and contig of chromosome segment 11p15.5 and genes in the LOH11A metastasis suppressor region. Genomics. 55:164–175. - PubMed
-
- Boyle, S., S. Gilchrist, J.M. Bridger, N.L. Mahy, J.A. Ellis, and W.A. Bickmore. 2001. The spatial organization of human chromosomes in the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219. - PubMed
-
- Brown, K.E., S. Amoils, J.M. Horn, V.J. Buckle, D.R. Higgs, M. Merkenschlager, and A.G. Fisher. 2001. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3:602–606. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

