The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint
- PMID: 12473689
- PMCID: PMC2173375
- DOI: 10.1083/jcb.200205068
The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint
Abstract
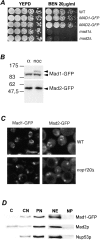
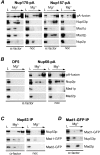
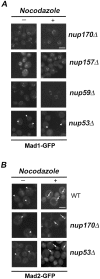
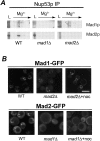
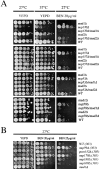
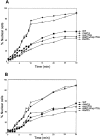
Aphysical and functional link between the nuclear pore complex (NPC) and the spindle checkpoint machinery has been established in the yeast Saccharomyces cerevisiae. We show that two proteins required for the execution of the spindle checkpoint, Mad1p and Mad2p, reside predominantly at the NPC throughout the cell cycle. There they are associated with a subcomplex of nucleoporins containing Nup53p, Nup170p, and Nup157p. The association of the Mad1p-Mad2p complex with the NPC requires Mad1p and is mediated in part by Nup53p. On activation of the spindle checkpoint, we detect changes in the interactions between these proteins, including the release of Mad2p (but not Mad1p) from the NPC and the accumulation of Mad2p at kinetochores. Accompanying these events is the Nup53p-dependent hyperphosphorylation of Mad1p. On the basis of these results and genetic analysis of double mutants, we propose a model in which Mad1p bound to a Nup53p-containing complex sequesters Mad2p at the NPC until its release by activation of the spindle checkpoint. Furthermore, we show that the association of Mad1p with the NPC is not passive and that it plays a role in nuclear transport.
Figures




References
-
- Adams, A., D.E. Gottschling, C.A. Kaiser, and T. Stearns. 1997. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 177 pp.
-
- Aitchison, J.D., M.P. Rout, M.M. Marelli, G. Blobel, and R.W. Wozniak. 1995. b. Two novel related yeast nucleoporins Nup170p and Nup157p: Complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131:1133–1148. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

