The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP
- PMID: 12473690
- PMCID: PMC2173378
- DOI: 10.1083/jcb.200208017
The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP
Abstract
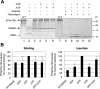
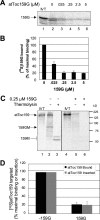
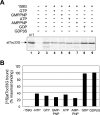
The multimeric translocon at the outer envelope membrane of chloroplasts (Toc) initiates the recognition and import of nuclear-encoded preproteins into chloroplasts. Two Toc GTPases, Toc159 and Toc33/34, mediate preprotein recognition and regulate preprotein translocation. Although these two proteins account for the requirement of GTP hydrolysis for import, the functional significance of GTP binding and hydrolysis by either GTPase has not been defined. A recent study indicates that Toc159 is equally distributed between a soluble cytoplasmic form and a membrane-inserted form, raising the possibility that it might cycle between the cytoplasm and chloroplast as a soluble preprotein receptor. In the present study, we examined the mechanism of targeting and insertion of the Arabidopsis thaliana orthologue of Toc159, atToc159, to chloroplasts. Targeting of atToc159 to the outer envelope membrane is strictly dependent only on guanine nucleotides. Although GTP is not required for initial binding, the productive insertion and assembly of atToc159 into the Toc complex requires its intrinsic GTPase activity. Targeting is mediated by direct binding between the GTPase domain of atToc159 and the homologous GTPase domain of atToc33, the Arabidopsis Toc33/34 orthologue. Our findings demonstrate a role for the coordinate action of the Toc GTPases in assembly of the functional Toc complex at the chloroplast outer envelope membrane.
Figures








Similar articles
-
Essential role of the G-domain in targeting of the protein import receptor atToc159 to the chloroplast outer membrane.J Cell Biol. 2002 Dec 9;159(5):845-54. doi: 10.1083/jcb.200208018. Epub 2002 Dec 2. J Cell Biol. 2002. PMID: 12460988 Free PMC article.
-
The roles of toc34 and toc75 in targeting the toc159 preprotein receptor to chloroplasts.J Biol Chem. 2003 Nov 7;278(45):44289-97. doi: 10.1074/jbc.M307873200. Epub 2003 Sep 1. J Biol Chem. 2003. PMID: 12951325
-
A toc159 import receptor mutant, defective in hydrolysis of GTP, supports preprotein import into chloroplasts.J Biol Chem. 2009 Mar 27;284(13):8670-9. doi: 10.1074/jbc.M804235200. Epub 2009 Feb 2. J Biol Chem. 2009. PMID: 19188370 Free PMC article.
-
Protein transport in organelles: The Toc complex way of preprotein import.FEBS J. 2009 Mar;276(5):1156-65. doi: 10.1111/j.1742-4658.2009.06873.x. FEBS J. 2009. PMID: 19187236 Review.
-
The gateway to chloroplast: re-defining the function of chloroplast receptor proteins.Biol Chem. 2012 Nov;393(11):1263-77. doi: 10.1515/hsz-2012-0235. Biol Chem. 2012. PMID: 23109543 Review.
Cited by
-
The acidic domains of the Toc159 chloroplast preprotein receptor family are intrinsically disordered protein domains.BMC Biochem. 2009 Dec 30;10:35. doi: 10.1186/1471-2091-10-35. BMC Biochem. 2009. PMID: 20042108 Free PMC article.
-
AtToc90, a new GTP-binding component of the Arabidopsis chloroplast protein import machinery.Plant Mol Biol. 2004 Feb;54(3):427-40. doi: 10.1023/B:PLAN.0000036374.92546.51. Plant Mol Biol. 2004. PMID: 15284497
-
Split-Ubiquitin Two-Hybrid Screen for Proteins Interacting with slToc159-1 and slToc159-2, Two Chloroplast Preprotein Import Receptors in Tomato (Solanum lycopersicum).Plants (Basel). 2022 Oct 30;11(21):2923. doi: 10.3390/plants11212923. Plants (Basel). 2022. PMID: 36365376 Free PMC article.
-
Molecular characterization, targeting and expression analysis of chloroplast and mitochondrion protein import components in Nicotiana benthamiana.Front Plant Sci. 2022 Oct 26;13:1040688. doi: 10.3389/fpls.2022.1040688. eCollection 2022. Front Plant Sci. 2022. PMID: 36388587 Free PMC article.
-
The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance.Front Physiol. 2015 Sep 24;6:259. doi: 10.3389/fphys.2015.00259. eCollection 2015. Front Physiol. 2015. PMID: 26441678 Free PMC article. Review.
References
-
- Bauer, J., K. Chen, A. Hiltbunner, E. Wehrli, M. Eugster, D. Schnell, and F. Kessler. 2000. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 403:203–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

