Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts
- PMID: 12475934
- PMCID: PMC139199
- DOI: 10.1073/pnas.262591699
Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts
Abstract
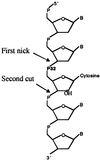
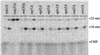
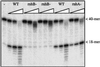
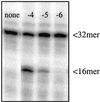
Misincorporated ribonucleotides in DNA will cause DNA backbone distortion and may be targeted by DNA repair enzymes. Using double-stranded oligonucleotide probes containing a single ribose, we demonstrate a robust activity in human, yeast, and Escherichia coli cell-free extracts that nicks 5' of the ribose. The human and yeast extracts also make a subsequent cut 3' of the ribonucleotide releasing a ribonucleotide monophosphate. The resulting 1-nt gap is an ideal substrate for polymerase and ligase to complete a proposed repair sequence that effectively replaces the ribose with deoxyribose. Screening of yeast deletion mutant cells reveals that the initial nick is made by RNase H(35), a RNase H type 2 enzyme, and the second cut is made by Rad27p, the yeast homologue of human FEN-1 protein. RNase H type 2 enzymes are present in all kingdoms of life and are evolutionarily well conserved. We knocked out the corresponding rnhb gene in E. coli and show that extracts from this strain lack the nicking activity. Conversely, a highly purified archaeal RNase HII type 2 protein has a pronounced activity. To study substrate specificity, extracts were made from a yeast double mutant lacking the other main RNase H enzymes [RNase H1 and RNase H(70)], while maintaining RNase H(35). It was found that a single ribose is preferred as substrate over a stretch of riboses, further strengthening a proposed role of this enzyme in the repair of misincorporated ribonucleotides rather than (or in addition to) processing RNADNA hybrid molecules.
Figures





References
-
- von Sonntag C. & Schulte-Frohlinde, D. (1978) Mol. Biol. Biochem. Biophys. 27, 204-226. - PubMed
-
- Isildar M., Schuchmann, M. N., Schulte-Frohlinde, D. & von Sonntag, C. (1981) Int. J. Radiat. Biol. 40, 347-354. - PubMed
-
- von Sonntag C. (1984) Int. J. Radiat. Biol. 46, 507-519. - PubMed
-
- Horton N. C. & Finzel, B. C. (1996) J. Mol. Biol. 264, 521-533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

