Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos
- PMID: 12475958
- PMCID: PMC138639
- DOI: 10.1091/mbc.e02-06-0346
Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos
Erratum in
- Mol Biol Cell. 2004 Jan;15(1):preceding Table of Contents
Abstract
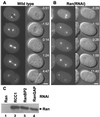
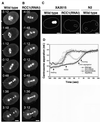
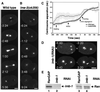
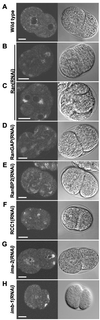
The small GTPase Ran has been found to play pivotal roles in several aspects of cell function. We have investigated the role of the Ran GTPase cycle in spindle formation and nuclear envelope assembly in dividing Caenorhabditis elegans embryos in real time. We found that Ran and its cofactors RanBP2, RanGAP, and RCC1 are all essential for reformation of the nuclear envelope after cell division. Reducing the expression of any of these components of the Ran GTPase cycle by RNAi leads to strong extranuclear clustering of integral nuclear envelope proteins and nucleoporins. Ran, RanBP2, and RanGAP are also required for building a mitotic spindle, whereas astral microtubules are normal in the absence of these proteins. RCC1(RNAi) embryos have similar abnormalities in the initial phase of spindle formation but eventually recover to form a bipolar spindle. Irregular chromatin structures and chromatin bridges due to spindle failure were frequently observed in embryos where the Ran cycle was perturbed. In addition, connection between the centrosomes and the male pronucleus, and thus centrosome positioning, depends upon the Ran cycle components. Finally, we have demonstrated that both IMA-2 and IMB-1, the homologues of vertebrate importin alpha and beta, are essential for both spindle assembly and nuclear formation in early embryos.
Figures




References
-
- Bamba C, Bobinnec Y, Fukuda M, Nishida E. The GTPase Ran regulates chromosome positioning and nuclear envelope assembly in vivo. Curr Biol. 2002;12:503–507. - PubMed
-
- Bilbao-Cortés D, Hetzer M, Laengst G, Becker PB, Mattaj IW. Ran binds to chromatin by two distinct mechanisms. Curr Biol. 2002;12:1151–1156. - PubMed
-
- Bischoff FR, Görlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. - PubMed
-
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

