Screening of transgenic proteins expressed in transgenic food crops for the presence of short amino acid sequences identical to potential, IgE - binding linear epitopes of allergens
- PMID: 12477382
- PMCID: PMC139984
- DOI: 10.1186/1472-6807-2-8
Screening of transgenic proteins expressed in transgenic food crops for the presence of short amino acid sequences identical to potential, IgE - binding linear epitopes of allergens
Abstract
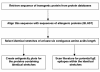
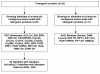
Background: Transgenic proteins expressed by genetically modified food crops are evaluated for their potential allergenic properties prior to marketing, among others by identification of short identical amino acid sequences that occur both in the transgenic protein and allergenic proteins. A strategy is proposed, in which the positive outcomes of the sequence comparison with a minimal length of six amino acids are further screened for the presence of potential linear IgE-epitopes. This double track approach involves the use of literature data on IgE-epitopes and an antigenicity prediction algorithm.
Results: Thirty-three transgenic proteins have been screened for identities of at least six contiguous amino acids shared with allergenic proteins. Twenty-two transgenic proteins showed positive results of six- or seven-contiguous amino acids length. Only a limited number of identical stretches shared by transgenic proteins (papaya ringspot virus coat protein, acetolactate synthase GH50, and glyphosate oxidoreductase) and allergenic proteins could be identified as (part of) potential linear epitopes.
Conclusion: Many transgenic proteins have identical stretches of six or seven amino acids in common with allergenic proteins. Most identical stretches are likely to be false positives. As shown in this study, identical stretches can be further screened for relevance by comparison with linear IgE-binding epitopes described in literature. In the absence of literature data on epitopes, antigenicity prediction by computer aids to select potential antibody binding sites that will need verification of IgE binding by sera binding tests. Finally, the positive outcomes of this approach warrant further clinical testing for potential allergenicity.
Figures


Similar articles
-
Presence of potential allergy-related linear epitopes in novel proteins from conventional crops and the implication for the safety assessment of these crops with respect to the current testing of genetically modified crops.Plant Biotechnol J. 2003 Sep;1(5):371-80. doi: 10.1046/j.1467-7652.2003.00035.x. Plant Biotechnol J. 2003. PMID: 17166136
-
Molecular basis of IgE-recognition of Lol p 5, a major allergen of rye-grass pollen.Mol Immunol. 1998 Apr;35(5):293-305. doi: 10.1016/s0161-5890(98)00050-9. Mol Immunol. 1998. PMID: 9747889
-
Lack of cross-reactivity between the Bacillus thuringiensis derived protein Cry1F in maize grain and dust mite Der p7 protein with human sera positive for Der p7-IgE.Regul Toxicol Pharmacol. 2006 Mar;44(2):136-43. doi: 10.1016/j.yrtph.2005.11.005. Epub 2006 Jan 9. Regul Toxicol Pharmacol. 2006. PMID: 16406630
-
Will genetically modified foods be allergenic?J Allergy Clin Immunol. 2001 May;107(5):765-71. doi: 10.1067/mai.2001.114241. J Allergy Clin Immunol. 2001. PMID: 11344340 Review.
-
The structural requirements of epitopes with IgE binding capacity demonstrated by three major allergens from fish, egg and tree pollen.Scand J Clin Lab Invest Suppl. 1991;204:17-31. Scand J Clin Lab Invest Suppl. 1991. PMID: 1710368 Review.
Cited by
-
EVALLER: a web server for in silico assessment of potential protein allergenicity.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W694-700. doi: 10.1093/nar/gkm370. Epub 2007 May 30. Nucleic Acids Res. 2007. PMID: 17537818 Free PMC article.
-
Allermatch, a webtool for the prediction of potential allergenicity according to current FAO/WHO Codex alimentarius guidelines.BMC Bioinformatics. 2004 Sep 16;5:133. doi: 10.1186/1471-2105-5-133. BMC Bioinformatics. 2004. PMID: 15373946 Free PMC article.
-
PREAL: prediction of allergenic protein by maximum Relevance Minimum Redundancy (mRMR) feature selection.BMC Syst Biol. 2013;7 Suppl 5(Suppl 5):S9. doi: 10.1186/1752-0509-7-S5-S9. Epub 2013 Dec 9. BMC Syst Biol. 2013. PMID: 24565053 Free PMC article.
-
Arabitol dehydrogenase as a selectable marker for rice.Plant Cell Rep. 2005 Dec;24(10):596-602. doi: 10.1007/s00299-005-0015-3. Epub 2005 Nov 16. Plant Cell Rep. 2005. PMID: 16151815
-
AlgPred: prediction of allergenic proteins and mapping of IgE epitopes.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W202-9. doi: 10.1093/nar/gkl343. Nucleic Acids Res. 2006. PMID: 16844994 Free PMC article.
References
-
- James C. Global Review of Commercialized Transgenic Crops: 2001. Ithaca, International Service for the Acquisition of Agri-biotech Applications. 2001. http://www.isaaa.org/publications/briefs/Brief_24.htm
-
- Metcalfe DD, Astwood JD, Townsend R, Sampson HA, Taylor SL, Fuchs RL. Assessment of the allergenic potential of foods derived from genetically engineered crop plants. Crit Rev Food Sci Nutr. 1996;36:S165–S186. - PubMed
-
- FAO/WHO Draft Guideline for the Conduct of Food Safety Assessment of Foods Derived from Recombinant-DNA Plants (ALINORM 01/34A). Rome, Codex Alimentarius Committee, Food and Agriculture Organisation of the United Nations. 2002. http://www.who.int/fsf/GMfood/codex_index.htm
-
- FAO/WHO Joint FAO/WHO Expert Consultation on Foods Derived from Biotechnology – Allergenicity of Genetically Modified Foods – Rome, 22 – 25 January 2001. Rome, Food and Agriculture Organisation of the United Nations. 2001. http://www.fao.org/es/esn/food/pdf/allergygm.pdf
-
- FAO/WHO Report of the Working Group 12 September 2001, Ad Hoc Open-Ended Working Group on Allergenicity, Vancouver, 10–12 September 2001, Hosted by the Government of Canada. Rome, Food and Agriculture Organisation of the United Nations. 2001. ftp://ftp.fao.org/codex/ccfbt3/wgreport_e.pdf
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

