The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis
- PMID: 12477831
- PMCID: PMC140594
- DOI: 10.1128/jvi.77.1.258-269.2003
The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis
Abstract
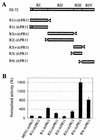
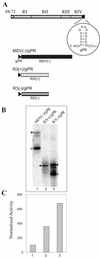
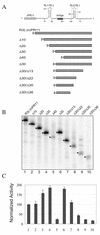
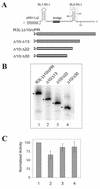
Replication of the RNA genomes of tombusviruses, which are small plus-sense RNA viruses of plants, may be regulated by cis-acting elements, including promoters and replication enhancers that are present in the RNA templates. Using a partially purified RNA-dependent RNA polymerase (RdRp) preparation (P. D. Nagy and J. Pogany, Virology 276:279-288, 2000), we demonstrate that the minus-strand templates of tombusviruses contain a replication enhancer, which can upregulate RNA synthesis initiating from the minimal plus-strand initiation promoter by 10- to 20-fold in an in vitro assay. Dissection of the sequence of the replication enhancer element revealed that the two stem-loop structures present within the approximately 80-nucleotide-long enhancer region have interchangeable roles in upregulating RNA synthesis. The single-stranded sequence located between the two stem-loops also plays an important role in stimulation of RNA synthesis. We also demonstrate that one of the two hairpins, both of which are similar to the hairpin of the minus-strand initiation promoter, can function as a promoter in vitro in the presence of short cytidylate-containing initiation sites. Overall, the in vitro data presented are consistent with previous in vivo results (D. Ray and K. A. White, Virology 256:162-171, 1999) and they firmly establish the presence of a replication enhancer on the minus-stranded RNA of tombusviruses.
Figures




Similar articles
-
Mechanism of stimulation of plus-strand synthesis by an RNA replication enhancer in a tombusvirus.J Virol. 2005 Aug;79(15):9777-85. doi: 10.1128/JVI.79.15.9777-9785.2005. J Virol. 2005. PMID: 16014939 Free PMC article.
-
Enhancement of RNA synthesis by promoter duplication in tombusviruses.Virology. 2003 May 25;310(1):118-29. doi: 10.1016/s0042-6822(03)00105-3. Virology. 2003. PMID: 12788636
-
Analysis of minimal promoter sequences for plus-strand synthesis by the Cucumber necrosis virus RNA-dependent RNA polymerase.Virology. 2002 May 10;296(2):263-74. doi: 10.1006/viro.2002.1423. Virology. 2002. PMID: 12069525
-
Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication.Virology. 2006 Jan 5;344(1):211-20. doi: 10.1016/j.virol.2005.09.017. Virology. 2006. PMID: 16364751 Review.
-
Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination.Prog Nucleic Acid Res Mol Biol. 2004;78:187-226. doi: 10.1016/S0079-6603(04)78005-8. Prog Nucleic Acid Res Mol Biol. 2004. PMID: 15210331 Review.
Cited by
-
The p92 polymerase coding region contains an internal RNA element required at an early step in Tombusvirus genome replication.J Virol. 2005 Apr;79(8):4848-58. doi: 10.1128/JVI.79.8.4848-4858.2005. J Virol. 2005. PMID: 15795270 Free PMC article.
-
A replication silencer element in a plus-strand RNA virus.EMBO J. 2003 Oct 15;22(20):5602-11. doi: 10.1093/emboj/cdg523. EMBO J. 2003. PMID: 14532132 Free PMC article.
-
An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs.J Virol. 2003 Jan;77(1):245-57. doi: 10.1128/jvi.77.1.245-257.2003. J Virol. 2003. PMID: 12477830 Free PMC article.
-
Conformational organization of the 3' untranslated region in the tomato bushy stunt virus genome.RNA. 2006 Dec;12(12):2199-210. doi: 10.1261/rna.238606. Epub 2006 Oct 31. RNA. 2006. PMID: 17077273 Free PMC article.
-
Defining the roles of cis-acting RNA elements in tombusvirus replicase assembly in vitro.J Virol. 2012 Jan;86(1):156-71. doi: 10.1128/JVI.00404-11. Epub 2011 Oct 19. J Virol. 2012. PMID: 22013057 Free PMC article.
References
-
- Axelrod, V. D., E. Brown, C. Priano, and D. R. Mills. 1991. Coliphage Q beta RNA replication: RNA catalytic for single-strand release. Virology 184:595-608. - PubMed
-
- Burgyan, J., L. Rubino, and M. Russo. 1991. De novo generation of cymbidium ringspot virus defective interfering RNA. J. Gen. Virol. 72:505-509. - PubMed
-
- Chang, Y. C., M. Borja, H. B. Scholthof, A. O. Jackson, and T. J. Morris. 1995. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology 210:41-53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

