Genome variability and capsid structural constraints of hepatitis a virus
- PMID: 12477850
- PMCID: PMC140588
- DOI: 10.1128/jvi.77.1.452-459.2003
Genome variability and capsid structural constraints of hepatitis a virus
Abstract
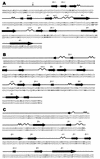
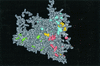
The number of synonymous mutations per synonymous site (K(s)), the number of nonsynonymous mutations per nonsynonymous site (K(a)), and the codon usage statistic (N(c)) were calculated for several hepatitis A virus (HAV) isolates. While K(s) was similar to those of poliovirus (PV) and foot-and-mouth disease virus (FMDV), K(a) was 1 order of magnitude lower. The N(c) parameter provides information on codon usage bias and decreases when bias increases. The N(c) value in HAV was about 38, while in PV and FMDV, it was about 53. The emergence of 22 rare codons in front of 8 in PV and 7 in FMDV was detected. Most of the conserved rare codons of the P1 region were strategically located at the carboxy borders of beta barrels and alpha helices, their potential function being the assurance of proper folding of the capsid proteins through a decrease in the translation speed. This strategic location was not observed for amino acids encoded by the conserved rare codons of the 3D region. The percentage of bases with low pairing number values was higher in the latter region, suggesting a role of the conserved rare codons in the maintenance of RNA structure. Many of the rare codons in HAV are among the most frequent in humans, unlike in PV or in FMDV. This fact may be explained by the lack of cellular shutoff in HAV. One hypothesis is that HAV has evolved in order to avoid competition with its host for cellular tRNAs.
Figures
References
-
- Acharya, R., E. Fry, D. Stuart, G. Fox, D. Rowlands, and F. Brown. 1989. The three dimensional structure of foot-and-mouth disease virus at 2.8 Å resolution. Nature 337:709-716. - PubMed
-
- Adzhubei, A. A., I. A. Adzhubei, I. A. Krasheninnikov, and S. Neidle. 1996. Non-random usage of “degenerate”codons is related to protein three-dimensional structure. FEBS Lett. 399:78-82. - PubMed
-
- Arauz-Ruiz, P., L. Sundqvist, Z. García, L. Taylor, K. Visoná, H. Norder, and L. O. Magnius. 2001. Presumed common source outbreaks of hepatitis A in an endemic area confirmed by limited sequencing within the VP1 region. J. Med. Virol. 65:449-456. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

