Trbp111 selectively binds a noncovalently assembled tRNA-like structure
- PMID: 12481025
- PMCID: PMC139195
- DOI: 10.1073/pnas.262667999
Trbp111 selectively binds a noncovalently assembled tRNA-like structure
Abstract
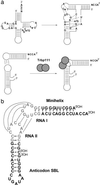
Transfer RNAs are key components of the genetic code by virtue of aminoacylation reactions whereby each amino acid is linked to the tRNA that bears the anticodon for the attached amino acid. The L-shaped tRNA structure contains two domains connected at right angles through a corner formed from tertiary interactions involving loops of each domain. Some evidence suggests that the domains arose separately and eventually were fused into a single covalent structure. In this scenario, the present-day tRNA possibly developed through a noncovalently assembled heterodimeric intermediate. Trbp111 is an ancient structure-specific tRNA binding protein that interacts specifically with the outside corner of the L-shaped molecule. Plausibly, this protein could act as a chaperone to cover and protect the fragile corner and thereby have a historical role in the development of tRNA. Here we show that Trbp111 interacts with a noncovalently assembled tRNA-like structure, under conditions where it does not interact with individual tRNA domains. Trbp111 binding specifically requires formation of the tRNA-like corner. In a mixture of RNA domains, it selects those that can make the L-like structure. Thus, cofactors such as Trbp111 have the capacity to help assemble and stabilize RNA dimers that are tRNA-like.
Figures







Similar articles
-
Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus.EMBO J. 1999 Jun 15;18(12):3475-83. doi: 10.1093/emboj/18.12.3475. EMBO J. 1999. PMID: 10369686 Free PMC article.
-
Crystal structure of trbp111: a structure-specific tRNA-binding protein.EMBO J. 2000 Dec 1;19(23):6287-98. doi: 10.1093/emboj/19.23.6287. EMBO J. 2000. PMID: 11101501 Free PMC article.
-
Simultaneous binding of two proteins to opposite sides of a single transfer RNA.Nat Struct Biol. 2001 Apr;8(4):344-8. doi: 10.1038/86228. Nat Struct Biol. 2001. PMID: 11276256
-
Aminoacylation of RNA minihelices: implications for tRNA synthetase structural design and evolution.Crit Rev Biochem Mol Biol. 1993;28(4):309-22. doi: 10.3109/10409239309078438. Crit Rev Biochem Mol Biol. 1993. PMID: 7691478 Review.
-
OB or Not OB: Idiosyncratic utilization of the tRNA-binding OB-fold domain in unicellular, pathogenic eukaryotes.FEBS Lett. 2016 Dec;590(23):4180-4191. doi: 10.1002/1873-3468.12441. Epub 2016 Oct 24. FEBS Lett. 2016. PMID: 27714804 Review.
Cited by
-
Transfer RNA Modification Enzymes from Thermophiles and Their Modified Nucleosides in tRNA.Microorganisms. 2018 Oct 20;6(4):110. doi: 10.3390/microorganisms6040110. Microorganisms. 2018. PMID: 30347855 Free PMC article. Review.
-
Bridging the gap between ribosomal and nonribosomal protein synthesis.Proc Natl Acad Sci U S A. 2010 Aug 17;107(33):14517-8. doi: 10.1073/pnas.1009939107. Epub 2010 Aug 9. Proc Natl Acad Sci U S A. 2010. PMID: 20696925 Free PMC article. No abstract available.
-
Combinatorial Fusion Rules to Describe Codon Assignment in the Standard Genetic Code.Life (Basel). 2020 Dec 23;11(1):4. doi: 10.3390/life11010004. Life (Basel). 2020. PMID: 33374866 Free PMC article.
-
RNA Aptamers for a tRNA-Binding Protein from Aeropyrum pernix with Homologous Counterparts Distributed Throughout Evolution.Life (Basel). 2020 Feb 1;10(2):11. doi: 10.3390/life10020011. Life (Basel). 2020. PMID: 32024042 Free PMC article.
-
Binding Properties of Split tRNA to the C-terminal Domain of Methionyl-tRNA Synthetase of Nanoarchaeum equitans.J Mol Evol. 2017 Jun;84(5-6):267-278. doi: 10.1007/s00239-017-9796-6. Epub 2017 Jun 6. J Mol Evol. 2017. PMID: 28589220
References
-
- Lapointe J. & Giegé, R. (1991) in Translation in Eukaryotes, ed. Trachsel, H. (CRC, Boca Raton, FL), pp. 35–69.
-
- Giegé R., Puglisi, J. D. & Florentz, C. (1993) Prog. Nucleic Acid Res. Mol. Biol. 45, 129-206. - PubMed
-
- Carter C. W. (1993) Annu. Rev. Biochem. 62, 715-748. - PubMed
-
- Noller H. F. (1993) in The RNA World, eds. Gesteland, R. F. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 137–156.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

