A structural model for Alzheimer's beta -amyloid fibrils based on experimental constraints from solid state NMR
- PMID: 12481027
- PMCID: PMC139214
- DOI: 10.1073/pnas.262663499
A structural model for Alzheimer's beta -amyloid fibrils based on experimental constraints from solid state NMR
Abstract
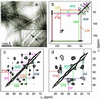
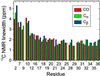
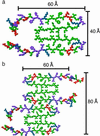
We present a structural model for amyloid fibrils formed by the 40-residue beta-amyloid peptide associated with Alzheimer's disease (Abeta(1-40)), based on a set of experimental constraints from solid state NMR spectroscopy. The model additionally incorporates the cross-beta structural motif established by x-ray fiber diffraction and satisfies constraints on Abeta(1-40) fibril dimensions and mass-per-length determined from electron microscopy. Approximately the first 10 residues of Abeta(1-40) are structurally disordered in the fibrils. Residues 12-24 and 30-40 adopt beta-strand conformations and form parallel beta-sheets through intermolecular hydrogen bonding. Residues 25-29 contain a bend of the peptide backbone that brings the two beta-sheets in contact through sidechain-sidechain interactions. A single cross-beta unit is then a double-layered beta-sheet structure with a hydrophobic core and one hydrophobic face. The only charged sidechains in the core are those of D23 and K28, which form salt bridges. Fibrils with minimum mass-per-length and diameter consist of two cross-beta units with their hydrophobic faces juxtaposed.
Figures


Comment in
-
Unraveling the secrets of Alzheimer's beta-amyloid fibrils.Proc Natl Acad Sci U S A. 2003 Jan 21;100(2):383-5. doi: 10.1073/pnas.0337745100. Epub 2003 Jan 13. Proc Natl Acad Sci U S A. 2003. PMID: 12525689 Free PMC article. No abstract available.
References
-
- Sipe J. D. (1992) Annu. Rev. Biochem. 61, 947-975. - PubMed
-
- Sunde M., Serpell, L. C., Bartlam, M., Fraser, P. E., Pepys, M. B. & Blake, C. C. F. (1997) J. Mol. Biol. 273, 729-739. - PubMed
-
- Glenner G. G. & Wong, C. W. (1984) Biochem. Biophys. Res. Commun. 120, 885-890. - PubMed
-
- Iwatsubo T., Odaka, A., Suzuki, N., Mizusawa, H., Nukina, N. & Ihara, Y. (1994) Neuron 13, 45-53. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

