Comparative molecular and functional analyses of the tobacco cyclin-dependent kinase inhibitor NtKIS1a and its spliced variant NtKIS1b
- PMID: 12481070
- PMCID: PMC166698
- DOI: 10.1104/pp.008573
Comparative molecular and functional analyses of the tobacco cyclin-dependent kinase inhibitor NtKIS1a and its spliced variant NtKIS1b
Abstract
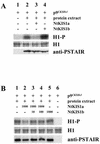
In all eukaryotes, cell cycle progression is controlled by cyclin-dependent kinases (CDKs) whose activity is regulated at several levels including inhibition by CDK inhibitors. Here, we report a comparative molecular and functional analysis of the tobacco (Nicotiana tomentosiformis) CDK inhibitor, NtKIS1a, and its spliced variant, NtKIS1b. The C-terminal end of NtKIS1a shares strong sequence similarity with mammalian CIP/KIP inhibitors, which is not the case for NtKIS1b. Consistent with this, NtKIS1a but not NtKIS1b inhibits in vitro the kinase activity of CDK/cyclin complexes, and tobacco (Nicotiana tabacum) D-type cyclins and an A-type CDK are NtKIS1a, but not NtKIS1b, interacting partners. Although both NtKIS1a and NtKIS1b transcripts are mainly found in flowers and more precisely in stamens, NtKIS1b transcript levels are cell cycle regulated, whereas those of NtKIS1a remain constant during the cell cycle. NtKIS1a and NtKIS1b fused to fluorescent proteins are localized in the nucleus when transiently expressed in onion epidermal cells. Furthermore, there is no competition for their nuclear localization when they are simultaneously overexpressed. In vitro competition toward CDK kinase activity suggests that NtKIS1b is a strong competitor of NtKIS1a. Arabidopsis plants overexpressing NtKIS1a-green fluorescent protein (GFP) or NtKIS1b-GFP fusion proteins were obtained. In these plants, the fusion proteins are still localized in the nucleus. Interestingly, NtKIS1a-GFP-overexpressing plants display strong morphological modifications and a reduced CDK kinase activity, whereas NtKIS1b-GFP-overexpressing plants display a wild-type phenotype including a wild-type CDK kinase activity. Our results strongly suggest that the inhibition of the kinase activity is responsible for the phenotypic modifications.
Figures




References
-
- Azzi L, Meijer L, Reed SI, Pidikiti R, Tung HY. Interaction between the cell-cycle-control proteins p34cdc2 and p9CKShs2: evidence for two cooperative binding domains in p9CKShs2. Eur J Biochem. 1992;203:353–360. - PubMed
-
- Brown JWS, Simpson CG. Splice site selection in plant pre-mRNA splicing. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:77–95. - PubMed
-
- Chen IT, Akamatsu M, Smith ML, Lung FD, Duba D, Roller PP, Fornace AJ, O'Connor PM. Characterization of p21Cip1/Waf1 peptide domains required for cyclin E/Cdk2 and PCNA interaction. Oncogene. 1996;12:595–607. - PubMed
-
- Davis SJ, Vierstra RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol. 1998;36:521–528. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

