Light-dependent death of maize lls1 cells is mediated by mature chloroplasts
- PMID: 12481072
- PMCID: PMC166700
- DOI: 10.1104/pp.008441
Light-dependent death of maize lls1 cells is mediated by mature chloroplasts
Abstract
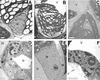
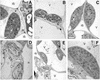
We reported previously the isolation of a novel cell death-suppressing gene from maize (Zea mays) encoded by the Lls1 (Lethal leaf spot-1) gene. Although the exact metabolic function of LLS1 remains elusive, here we provide insight into mechanisms that underlie the initiation and propagation of cell death associated with lls1 lesions. Our data indicate that lls1 lesions are triggered in response to a cell-damaging event caused by any biotic or abiotic agent or intrinsic metabolic imbalance--as long as the leaf tissue is developmentally competent to develop lls1 lesions. Continued expansion of these lesions, however, depends on the availability of light, with fluence rate being more important than spectral quality. Double-mutant analysis of lls1 with two maize mutants oil-yellow and iojap, both compromised photosynthetically and unable to accumulate normal levels of chlorophyll, indicated that it was the light harvested by the plant that energized lls1 lesion development. Chloroplasts appear to be the key mediators of lls1 cell death; their swelling and distortion occurs before any other changes normally associated with dying cells. In agreement with these results are indications that LLS1 is a chloroplast-localized protein whose transcript was detected only in green tissues. The propagative nature of light-dependent lls1 lesions predicts that cell death associated with these lesions is caused by a mobile agent such as reactive oxidative species. LLS1 may act to prevent reactive oxidative species formation or serve to remove a cell death mediator so as to maintain chloroplast integrity and cell survival.
Figures



References
-
- Aist JR, Gold RE, Bayles CJ, Morrison GH, Chandra S, Israel HW. Evidence that molecular components of papillae may be involved in ml-o resistance to barley powdery mildew. Physiol Mol Plant Pathol. 1988;33:17–32.
-
- Altschul SF, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons, Inc.; 1994.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

