Differential regulation of RNA levels of gibberellin dioxygenases by photoperiod in spinach
- PMID: 12481092
- PMCID: PMC166720
- DOI: 10.1104/pp.008581
Differential regulation of RNA levels of gibberellin dioxygenases by photoperiod in spinach
Abstract
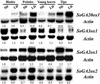
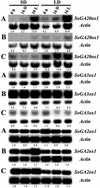
Previous work with spinach (Spinacia oleracea) has shown that the level of gibberellin (GA) 20-oxidase is strongly up-regulated by long days (LD). In the present work, the effect of photoperiod on expression of other GA dioxygenases was investigated and compared with that of GA 20-oxidase. Two GA 2-oxidases and one GA 3-oxidase were isolated from spinach by reverse transcription-polymerase chain reaction with degenerate primers and by 5'- and 3'-rapid amplification of cDNA ends. As determined by high-performance liquid chromatography with on-line radioactivity detection, the SoGA3ox1 gene product catalyzed 3beta-hydroxylation of GA(9) to GA(4) and GA(20) to GA(1). The SoGA2ox1 and the SoGA2ox2 gene products catalyzed 2beta-hydroxylation of GA(9) to GA(51) and GA(20) to GA(29). The product of GA(20) metabolism by SoGA3ox1 was identified as GA(1) by gas chromatography-mass spectrometry, whereas the products of GA(1) and GA(20) metabolism by SoGA2ox1 and SoGA2ox2 were identified as GA(8) and GA(29), respectively. SoGA2ox1 also metabolized GA(53) to GA(97). The levels of SoGA20ox1 transcripts were greatly increased in all organs tested in LD conditions, but the levels of SoGA3ox1 transcripts were only slightly increased in blades and petioles. A decrease in the levels of the SoGA2ox1 transcripts in young leaves and tips in LD conditions is opposite to the expression pattern of the SoGA20ox1. Expression of SoGA20ox1 in petioles and young leaves was strongly up-regulated by a supplementary 16 h of light, but the levels of SoGA3ox1 and SoGA2ox1 transcripts did not change. It is concluded that regulation and maintenance of GA(1) concentration in spinach are primarily attributable to changes in expression of SoGA20ox1.
Figures




Similar articles
-
Regulation of gibberellin 20-oxidase1 expression in spinach by photoperiod.Planta. 2007 Jun;226(1):35-44. doi: 10.1007/s00425-006-0463-1. Epub 2007 Jan 11. Planta. 2007. PMID: 17216482
-
Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach.Plant Physiol. 1996 Feb;110(2):547-54. doi: 10.1104/pp.110.2.547. Plant Physiol. 1996. PMID: 8742334 Free PMC article.
-
Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris.Plant Physiol. 2005 May;138(1):243-54. doi: 10.1104/pp.104.056499. Epub 2005 Apr 8. Plant Physiol. 2005. PMID: 15821147 Free PMC article.
-
Daylength and spatial expression of a gibberellin 20-oxidase isolated from hybrid aspen (Populus tremula L. x P. tremuloides Michx.).Planta. 2002 Apr;214(6):920-30. doi: 10.1007/s00425-001-0703-3. Epub 2001 Dec 15. Planta. 2002. PMID: 11941469
-
The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression.Proc Natl Acad Sci U S A. 1995 Jul 3;92(14):6640-4. doi: 10.1073/pnas.92.14.6640. Proc Natl Acad Sci U S A. 1995. PMID: 7604047 Free PMC article.
Cited by
-
Distribution and prediction of catalytic domains in 2-oxoglutarate dependent dioxygenases.BMC Res Notes. 2012 Aug 4;5:410. doi: 10.1186/1756-0500-5-410. BMC Res Notes. 2012. PMID: 22862831 Free PMC article.
-
Immunomodulation of gibberellin biosynthesis using an anti-precursor gibberellin antibody confers gibberellin-deficient phenotypes.Planta. 2008 Oct;228(5):863-73. doi: 10.1007/s00425-008-0788-z. Epub 2008 Jul 18. Planta. 2008. PMID: 18636270
-
Genome-Wide Analysis of the Biosynthesis and Deactivation of Gibberellin-Dioxygenases Gene Family in Camellia sinensis (L.) O. Kuntze.Genes (Basel). 2017 Sep 19;8(9):235. doi: 10.3390/genes8090235. Genes (Basel). 2017. PMID: 28925957 Free PMC article.
-
Divergence and adaptive evolution of the gibberellin oxidase genes in plants.BMC Evol Biol. 2015 Sep 29;15:207. doi: 10.1186/s12862-015-0490-2. BMC Evol Biol. 2015. PMID: 26416509 Free PMC article.
-
Diurnal regulation of the brassinosteroid-biosynthetic CPD gene in Arabidopsis.Plant Physiol. 2006 May;141(1):299-309. doi: 10.1104/pp.106.079145. Epub 2006 Mar 10. Plant Physiol. 2006. PMID: 16531479 Free PMC article.
References
-
- Elliott RC, Ross JJ, Smith JJ, Lester DR, Reid JB. Feed-forward regulation of gibberellin deactivation in pea. J Plant Growth Regul. 2001;20:87–94.
-
- Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

