Cloning of beta-primeverosidase from tea leaves, a key enzyme in tea aroma formation
- PMID: 12481100
- PMCID: PMC166728
- DOI: 10.1104/pp.102.011023
Cloning of beta-primeverosidase from tea leaves, a key enzyme in tea aroma formation
Abstract
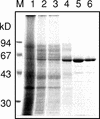
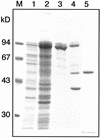
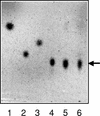
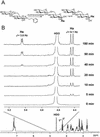
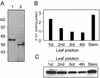
A beta-primeverosidase from tea (Camellia sinensis) plants is a unique disaccharide-specific glycosidase, which hydrolyzes aroma precursors of beta-primeverosides (6-O-beta-D-xylopyranosyl-beta-D-glucopyranosides) to liberate various aroma compounds, and the enzyme is deeply concerned with the floral aroma formation in oolong tea and black tea during the manufacturing process. The beta-primeverosidase was purified from fresh leaves of a cultivar for green tea (C. sinensis var sinensis cv Yabukita), and its partial amino acid sequences were determined. The beta-primeverosidase cDNA has been isolated from a cDNA library of cv Yabukita using degenerate oligonucleotide primers. The cDNA insert encodes a polypeptide consisting of an N-terminal signal peptide of 28 amino acid residues and a 479-amino acid mature protein. The beta-primeverosidase protein sequence was 50% to 60% identical to beta-glucosidases from various plants and was classified in a family 1 glycosyl hydrolase. The mature form of the beta-primeverosidase expressed in Escherichia coli was able to hydrolyze beta-primeverosides to liberate a primeverose unit and aglycons, but did not act on 2-phenylethyl beta-D-glucopyranoside. These results indicate that the beta-primeverosidase selectively recognizes the beta-primeverosides as substrates and specifically hydrolyzes the beta-glycosidic bond between the disaccharide and the aglycons. The stereochemistry for enzymatic hydrolysis of 2-phenylethyl beta-primeveroside by the beta-primeverosidase was followed by (1)H-nuclear magnetic resonance spectroscopy, revealing that the enzyme hydrolyzes the beta-primeveroside by a retaining mechanism. The roles of the beta-primeverosidase in the defense mechanism in tea plants and the floral aroma formation during tea manufacturing process are also discussed.
Figures



Similar articles
-
Substrate specificity of beta-primeverosidase, a key enzyme in aroma formation during oolong tea and black tea manufacturing.Biosci Biotechnol Biochem. 2001 Dec;65(12):2719-29. doi: 10.1271/bbb.65.2719. Biosci Biotechnol Biochem. 2001. PMID: 11826969
-
Crystal structures of β-primeverosidase in complex with disaccharide amidine inhibitors.J Biol Chem. 2014 Jun 13;289(24):16826-34. doi: 10.1074/jbc.M114.553271. Epub 2014 Apr 21. J Biol Chem. 2014. PMID: 24753293 Free PMC article.
-
Furcatin hydrolase from Viburnum furcatum Blume is a novel disaccharide-specific acuminosidase in glycosyl hydrolase family 1.J Biol Chem. 2004 May 28;279(22):23405-14. doi: 10.1074/jbc.M311379200. Epub 2004 Feb 19. J Biol Chem. 2004. PMID: 14976214
-
Understanding the biosyntheses and stress response mechanisms of aroma compounds in tea (Camellia sinensis) to safely and effectively improve tea aroma.Crit Rev Food Sci Nutr. 2019;59(14):2321-2334. doi: 10.1080/10408398.2018.1506907. Epub 2018 Oct 2. Crit Rev Food Sci Nutr. 2019. PMID: 30277806 Review.
-
Rutinosidase and other diglycosidases: Rising stars in biotechnology.Biotechnol Adv. 2023 Nov;68:108217. doi: 10.1016/j.biotechadv.2023.108217. Epub 2023 Jul 20. Biotechnol Adv. 2023. PMID: 37481095 Review.
Cited by
-
Integrated RNA-Seq and sRNA-Seq Analysis Identifies Chilling and Freezing Responsive Key Molecular Players and Pathways in Tea Plant (Camellia sinensis).PLoS One. 2015 Apr 22;10(4):e0125031. doi: 10.1371/journal.pone.0125031. eCollection 2015. PLoS One. 2015. PMID: 25901577 Free PMC article.
-
Structural and enzymatic characterization of Os3BGlu6, a rice beta-glucosidase hydrolyzing hydrophobic glycosides and (1->3)- and (1->2)-linked disaccharides.Plant Physiol. 2009 Sep;151(1):47-58. doi: 10.1104/pp.109.139436. Epub 2009 Jul 8. Plant Physiol. 2009. PMID: 19587102 Free PMC article.
-
United States tea: A synopsis of ongoing tea research and solutions to United States tea production issues.Front Plant Sci. 2022 Sep 23;13:934651. doi: 10.3389/fpls.2022.934651. eCollection 2022. Front Plant Sci. 2022. PMID: 36212324 Free PMC article. Review.
-
The beta-glucosidases responsible for bioactivation of hydroxynitrile glucosides in Lotus japonicus.Plant Physiol. 2008 Jul;147(3):1072-91. doi: 10.1104/pp.107.109512. Epub 2008 May 8. Plant Physiol. 2008. PMID: 18467457 Free PMC article.
-
Functional characterization, homology modeling and docking studies of β-glucosidase responsible for bioactivation of cyanogenic hydroxynitrile glucosides from Leucaena leucocephala (subabul).Mol Biol Rep. 2013 Feb;40(2):1351-63. doi: 10.1007/s11033-012-2179-6. Epub 2012 Oct 19. Mol Biol Rep. 2013. PMID: 23079707
References
-
- Barrett T, Suresh CG, Tolley SP, Dodson EJ, Hughes MA. The crystal structure of a cyanogenic β-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure. 1995;3:951–960. - PubMed
-
- Bourbouze R, Pratviel-Sosa F, Percheron F. Rhamnodiastase and α-l-rhamnosidase de Fagopyrum esculentum. Phytochemistry. 1975;14:1279–1282.
-
- Bridel M. Primeverose, primeverosides and primeverosidase. C R Acad Sci Paris. 1925;180:1421–1425.
-
- Chassagne D, Crouzet JC, Bayonove CL, Brillouet JM, Baumes RL. 6-O-α-Arabinopyranosyl-β-d-glucopyranosides as aroma precursors from passion fruit. Phytochemistry. 1996;41:1497–1500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

