Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms
- PMID: 12481102
- PMCID: PMC166730
- DOI: 10.1104/pp.009654
Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms
Abstract
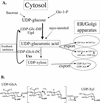
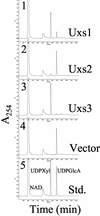
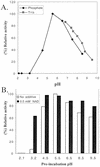
UDP-xylose (Xyl) is an important sugar donor for the synthesis of glycoproteins, polysaccharides, various metabolites, and oligosaccharides in animals, plants, fungi, and bacteria. UDP-Xyl also feedback inhibits upstream enzymes (UDP-glucose [Glc] dehydrogenase, UDP-Glc pyrophosphorylase, and UDP-GlcA decarboxylase) and is involved in its own synthesis and the synthesis of UDP-arabinose. In plants, biosynthesis of UDP-Xyl is catalyzed by different membrane-bound and soluble UDP-GlcA decarboxylase (UDP-GlcA-DC) isozymes, all of which convert UDP-GlcA to UDP-Xyl. Because synthesis of UDP-Xyl occurs both in the cytosol and in membranes, it is not known which source of UDP-Xyl the different Golgi-localized xylosyltransferases are utilizing. Here, we describe the identification of several distinct Arabidopsis genes (named AtUXS for UDP-Xyl synthase) that encode functional UDP-GlcA-DC isoforms. The Arabidopsis genome contains five UXS genes and their protein products can be subdivided into three isozyme classes (A-C), one soluble and two distinct putative membrane bound. AtUxs from each class, when expressed in Escherichia coli, generate active UDP-GlcA-DC that converts UDP-GlcA to UDP-Xyl. Members of this gene family have a large conserved C-terminal catalytic domain (approximately 300 amino acids long) and an N-terminal variable domain differing in sequence and size (30-120 amino acids long). Isoforms of class A and B appear to encode putative type II membrane proteins with their catalytic domains facing the lumen (like Golgi-glycosyltransferases) and their N-terminal variable domain facing the cytosol. Uxs class C is likely a cytosolic isoform. The characteristics of the plant Uxs support the hypothesis that unique UDP-GlcA-DCs with distinct subcellular localizations are required for specific xylosylation events.
Figures




References
-
- Amino SI, Takeuchi Y, Kamamine A. Changes in enzyme activities involved in the formation and interconversion of UDP-sugars during cell cycle in a synchronous culture of Catharanthus roseus. Physiol Plant. 1985;64:111–117.
-
- Ankel H, Ankel E, Feingold DS, Schutzbach JS. Formation of UDP-d-xylose in algae. Biochim Biophys Acta. 1967;136:172–175. - PubMed
-
- Ankel H, Feingold DS. Biosynthesis of uridine diphosphate d-xylose: I. Uridine diphosphate glucuronate carboxy-lyase of wheat germ. Biochemistry. 1965;4:2468–2475. - PubMed
-
- Ankel H, Feingold DS. Biosynthesis of uridine diphosphate d-xylose: II. Uridine diphosphate d-glucuronate carboxy-lyase of Cryptococcus laurentii. Biochemistry. 1966;5:182–189. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

