The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis
- PMID: 12482959
- PMCID: PMC140671
- DOI: 10.1128/MCB.23.1.38-54.2003
The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis
Abstract
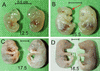
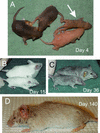
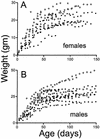
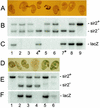
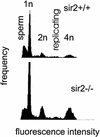
The yeast Sir2p protein has an essential role in maintaining telomeric and mating type genes in their transcriptionally inactive state. Mammalian cells have a very large proportion of their genome inactive and also contain seven genes that have regions of homology with the yeast sir2 gene. One of these mammalian genes, sir2alpha, is the presumptive mammalian homologue of the yeast sir2 gene. We set out to determine if sir2alpha plays a role in mammalian gene silencing by creating a strain of mice carrying a null allele of sir2alpha. Animals carrying two null alleles of sir2alpha were smaller than normal at birth, and most died during the early postnatal period. In an outbred background, the sir2alpha null animals often survived to adulthood, but both sexes were sterile. We found no evidence for failure of gene silencing in sir2alpha null animals, suggesting that either SIR2alpha has a different role in mammals than it does in Saccharomyces cerevisiae or that its role in gene silencing in confined to a small subset of mammalian genes. The phenotype of the sir2alpha null animals suggests that the SIR2alpha protein is essential for normal embryogenesis and for normal reproduction in both sexes.
Figures
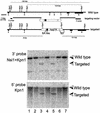
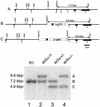
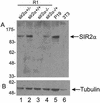
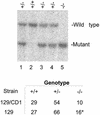


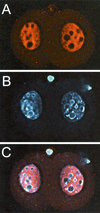
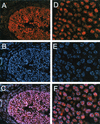
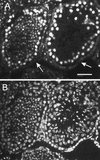
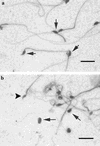



References
-
- Accili, D., J. Drago, E. J. Lee, M. D. Johnson, M. H. Cool, P. Salvatore, L. D. Asico, P. A. Jose, S. I. Taylor, and H. Westphal. 1996. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat. Genet. 12:106-109. - PubMed
-
- Adra, C. N., P. H. Boer, and M. W. McBurney. 1987. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene 60:65-74. - PubMed
-
- Adra, C. N., N. A. Ellis, and M. W. McBurney. 1988. The family of mouse phosphoglycerate kinase genes and pseudogenes. Somat. Cell Mol. Genet. 14:69-81. - PubMed
-
- Allemand, I., A. Anglo, A. Y. Jeantet, I. Cerutti, and E. May. 1999. Testicular wild-type p53 expression in transgenic mice induces spermiogenesis alterations ranging from differentiation defects to apoptosis. Oncogene 18:6521-6530. - PubMed
-
- Baker, J., J. P. Liu, E. J. Robertson, and A. Efstratiadis. 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75:73-82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
