An Rpb4/Rpb7-like complex in yeast RNA polymerase III contains the orthologue of mammalian CGRP-RCP
- PMID: 12482973
- PMCID: PMC140662
- DOI: 10.1128/MCB.23.1.195-205.2003
An Rpb4/Rpb7-like complex in yeast RNA polymerase III contains the orthologue of mammalian CGRP-RCP
Abstract
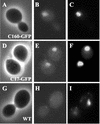
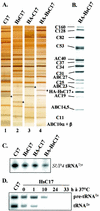
The essential C17 subunit of yeast RNA polymerase (Pol) III interacts with Brf1, a component of TFIIIB, suggesting a role for C17 in the initiation step of transcription. The protein sequence of C17 (encoded by RPC17) is conserved from yeasts to humans. However, mammalian homologues of C17 (named CGRP-RCP) are known to be involved in a signal transduction pathway related to G protein-coupled receptors, not in transcription. In the present work, we first establish that human CGRP-RCP is the genuine orthologue of C17. CGRP-RCP was found to functionally replace C17 in Deltarpc17 yeast cells; the purified mutant Pol III contained CGRP-RCP and had a decreased specific activity but initiated faithfully. Furthermore, CGRP-RCP was identified by mass spectrometry in a highly purified human Pol III preparation. These results suggest that CGRP-RCP has a dual function in mammals. Next, we demonstrate by genetic and biochemical approaches that C17 forms with C25 (encoded by RPC25) a heterodimer akin to Rpb4/Rpb7 in Pol II. C17 and C25 were found to interact genetically in suppression screens and physically in coimmunopurification and two-hybrid experiments. Sequence analysis and molecular modeling indicated that the C17/C25 heterodimer likely adopts a structure similar to that of the archaeal RpoE/RpoF counterpart of the Rpb4/Rpb7 complex. These RNA polymerase subunits appear to have evolved to meet the distinct requirements of the multiple forms of RNA polymerases.
Figures





Similar articles
-
Loss of the Rpb4/Rpb7 subcomplex in a mutant form of the Rpb6 subunit shared by RNA polymerases I, II, and III.Mol Cell Biol. 2003 May;23(9):3329-38. doi: 10.1128/MCB.23.9.3329-3338.2003. Mol Cell Biol. 2003. PMID: 12697831 Free PMC article.
-
A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70.Mol Cell Biol. 2000 Jan;20(2):488-95. doi: 10.1128/MCB.20.2.488-495.2000. Mol Cell Biol. 2000. PMID: 10611227 Free PMC article.
-
Rpb7 subunit of RNA polymerase II interacts with an RNA-binding protein involved in processing of transcripts.Nucleic Acids Res. 2003 Aug 15;31(16):4696-701. doi: 10.1093/nar/gkg688. Nucleic Acids Res. 2003. PMID: 12907709 Free PMC article.
-
Genetics of eukaryotic RNA polymerases I, II, and III.Microbiol Rev. 1993 Sep;57(3):703-24. doi: 10.1128/mr.57.3.703-724.1993. Microbiol Rev. 1993. PMID: 8246845 Free PMC article. Review.
-
Rpb4 and Rpb7: subunits of RNA polymerase II and beyond.Trends Biochem Sci. 2004 Dec;29(12):674-81. doi: 10.1016/j.tibs.2004.10.007. Trends Biochem Sci. 2004. PMID: 15544954 Review.
Cited by
-
Ancient origin, functional conservation and fast evolution of DNA-dependent RNA polymerase III.Nucleic Acids Res. 2006 Jul 28;34(13):3615-24. doi: 10.1093/nar/gkl421. Print 2006. Nucleic Acids Res. 2006. PMID: 16877568 Free PMC article.
-
Genomewide recruitment analysis of Rpb4, a subunit of polymerase II in Saccharomyces cerevisiae, reveals its involvement in transcription elongation.Eukaryot Cell. 2008 Jun;7(6):1009-18. doi: 10.1128/EC.00057-08. Epub 2008 Apr 25. Eukaryot Cell. 2008. PMID: 18441121 Free PMC article.
-
Crystallization of RNA polymerase I subcomplex A14/A43 by iterative prediction, probing and removal of flexible regions.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 May 1;64(Pt 5):413-8. doi: 10.1107/S174430910800972X. Epub 2008 Apr 24. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18453714 Free PMC article.
-
Skin-bacteria communication: Involvement of the neurohormone Calcitonin Gene Related Peptide (CGRP) in the regulation of Staphylococcus epidermidis virulence.Sci Rep. 2016 Oct 14;6:35379. doi: 10.1038/srep35379. Sci Rep. 2016. PMID: 27739485 Free PMC article.
-
Domainal organization of the lower eukaryotic homologs of the yeast RNA polymerase II core subunit Rpb7 reflects functional conservation.Nucleic Acids Res. 2004 Jan 2;32(1):201-10. doi: 10.1093/nar/gkh163. Print 2004. Nucleic Acids Res. 2004. PMID: 14704357 Free PMC article.
References
-
- Andrau, J.-C., and M. Werner. 2001. B"-associated factor(s) involved in RNA polymerase III preinitiation complex formation and start-site selection. Eur. J. Biochem. 268:5167-5175. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Balkan, W., E. L. Oates, G. A. Howard, and B. A. Roos. 1999. Testes exhibit elevated expression of calcitonin gene-related peptide receptor component protein. Endocrinology 140:1459-1469. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
