Novel transcriptional potentiation of BETA2/NeuroD on the secretin gene promoter by the DNA-binding protein Finb/RREB-1
- PMID: 12482979
- PMCID: PMC140679
- DOI: 10.1128/MCB.23.1.259-271.2003
Novel transcriptional potentiation of BETA2/NeuroD on the secretin gene promoter by the DNA-binding protein Finb/RREB-1
Abstract
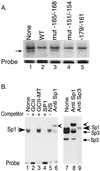
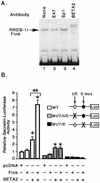
The basic helix-loop-helix protein BETA2/NeuroD activates transcription of the secretin gene and is essential for terminal differentiation of secretin-producing enteroendocrine cells. However, in heterodimeric complexes with its partner basic helix-loop-helix proteins, BETA2 does not appear to be a strong activator of transcription by itself. Mutational analysis of a proximal enhancer in the secretin gene identified several cis-acting elements in addition to the E-box binding site for BETA2. We identified by expression cloning the zinc finger protein RREB-1, also known to exist as a longer form, Finb, as the protein binding to one of the mutationally sensitive elements. Finb/RREB-1 lacks an intrinsic activation domain and by itself did not activate secretin gene transcription. Here we show that Finb/RREB-1 can associate with BETA2 to enhance its transcription-activating function. Both DNA binding and physical interaction of Finb/RREB-1 with BETA2 are required to potentiate transcription. Thus, Finb/RREB-1 does not function as a classical activator of transcription that recruits an activation domain to a DNA-protein complex. Finb/RREB-1 may be distinguished from coactivators, which increase transcription without sequence-specific DNA binding. We suggest that Finb/RREB-1 should be considered a potentiator of transcription, representing a distinct category of transcription-regulating proteins.
Figures

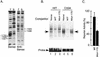





Similar articles
-
The basic helix-loop-helix transcription factor NeuroD1 facilitates interaction of Sp1 with the secretin gene enhancer.Mol Cell Biol. 2007 Nov;27(22):7839-47. doi: 10.1128/MCB.00438-07. Epub 2007 Sep 17. Mol Cell Biol. 2007. PMID: 17875929 Free PMC article.
-
p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene.Mol Cell Biol. 1998 May;18(5):2957-64. doi: 10.1128/MCB.18.5.2957. Mol Cell Biol. 1998. PMID: 9566915 Free PMC article.
-
The basic helix-loop-helix protein BETA2 interacts with p300 to coordinate differentiation of secretin-expressing enteroendocrine cells.Genes Dev. 1998 Mar 15;12(6):820-30. doi: 10.1101/gad.12.6.820. Genes Dev. 1998. PMID: 9512516 Free PMC article.
-
Review article: transcriptional events controlling the terminal differentiation of intestinal endocrine cells.Aliment Pharmacol Ther. 2000 Apr;14 Suppl 1:170-5. doi: 10.1046/j.1365-2036.2000.014s1170.x. Aliment Pharmacol Ther. 2000. PMID: 10807420 Review.
-
Transcriptional regulation of secretin gene expression.J Clin Gastroenterol. 1995;21 Suppl 1:S50-55. J Clin Gastroenterol. 1995. PMID: 8774991 Review.
Cited by
-
Embryonic nervous system genes predominate in searches for dinucleotide simple sequence repeats flanked by conserved sequences.Gene. 2009 Jan 15;429(1-2):74-9. doi: 10.1016/j.gene.2008.09.025. Epub 2008 Oct 2. Gene. 2009. PMID: 18952158 Free PMC article.
-
Functional Analysis of Two Zinc (Zn) Transporters (ZIP3 and ZIP8) Promoters and Their Distinct Response to MTF1 and RREB1 in the Regulation of Zn Metabolism.Int J Mol Sci. 2020 Aug 26;21(17):6135. doi: 10.3390/ijms21176135. Int J Mol Sci. 2020. PMID: 32858813 Free PMC article.
-
CtBP and associated LSD1 are required for transcriptional activation by NeuroD1 in gastrointestinal endocrine cells.Mol Cell Biol. 2014 Jun;34(12):2308-17. doi: 10.1128/MCB.01600-13. Epub 2014 Apr 14. Mol Cell Biol. 2014. PMID: 24732800 Free PMC article.
-
The retinoblastoma protein, RB, is required for gastrointestinal endocrine cells to exit the cell cycle, but not for hormone expression.Dev Biol. 2007 Nov 15;311(2):478-86. doi: 10.1016/j.ydbio.2007.08.052. Epub 2007 Sep 7. Dev Biol. 2007. PMID: 17936268 Free PMC article.
-
Loss of RREB1 reduces adipogenesis and improves insulin sensitivity in mouse and human adipocytes.bioRxiv [Preprint]. 2024 Jul 31:2024.07.30.605923. doi: 10.1101/2024.07.30.605923. bioRxiv. 2024. PMID: 39131393 Free PMC article. Preprint.
References
-
- Dumonteil, E., B. Laser, I. Constant, and J. Philippe. 1998. Differential regulation of the glucagon and insulin I gene promoters by basic helix-loop-helix transcription factors E47 and Beta2. J. Biol. Chem. 273:19945-19954. - PubMed
-
- Edlund, T., M. D. Walker, P. J. Barr, and W. J. Rutter. 1985. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5′ flanking elements. Science 230:912-916. - PubMed
-
- Fujimoto-Nishiyama, A., S. Ishii, S. Matsuda, J.-I. Inoue, and T. Yamamoto. 1997. A novel zinc finger protein, Finb, is a transcriptional activator and localized in nuclear bodies. Gene 195:267-275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
