Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor alpha-dependent transcription with chromatin templates
- PMID: 12482985
- PMCID: PMC140681
- DOI: 10.1128/MCB.23.1.335-348.2003
Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor alpha-dependent transcription with chromatin templates
Abstract
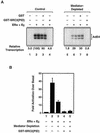
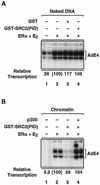
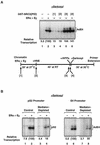
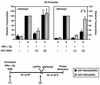
Ligand-dependent transcriptional activation by nuclear receptors involves the recruitment of various coactivators to the promoters of hormone-regulated genes assembled into chromatin. Nuclear receptor coactivators include histone acetyltransferase complexes, such as p300/CBP-steroid receptor coactivator (SRC), as well as the multisubunit mediator complexes ("Mediator"), which may help recruit RNA polymerase II to the promoter. We have used a biochemical approach, including an in vitro chromatin assembly and transcription system, to examine the functional role for Mediator in the transcriptional activity of estrogen receptor alpha (ERalpha) with chromatin templates, as well as functional interplay between Mediator and p300/CBP during ERalpha-dependent transcription. Using three different approaches to functionally inactivate Mediator (immunoneutralization, immunodepletion, and inhibitory polypeptides), we find that Mediator is required for maximal transcriptional activation by ligand-activated ERalpha. In addition, we demonstrate synergism between Mediator and p300/CBP-SRC during ERalpha-dependent transcription with chromatin templates, but not with naked DNA. This synergism is important for promoting the formation of a stable transcription preinitiation complex leading to the initiation of transcription. Interestingly, we find that Mediator has an additional distinct role during ERalpha-dependent transcription not shared by p300/CBP-SRC: namely, to promote preinitiation complex formation for subsequent rounds of transcription reinitiation. These results suggest that one functional consequence of Mediator-ERalpha interactions is the stimulation of multiple cycles of transcription reinitiation. Collectively, our results indicate an important role for Mediator, as well as its functional interplay with p300/CBP-SRC, in the enhancement of ERalpha-dependent transcription with chromatin templates.
Figures



References
-
- Boube, M., L. Joulia, D. L. Cribbs, and H. M. Bourbon. 2002. Evidence for a Mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. - PubMed
-
- Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. - PubMed
-
- Bulger, M., and J. T. Kadonaga. 1994. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol. Genet. 5:241-262.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
