Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation
- PMID: 12482986
- PMCID: PMC140685
- DOI: 10.1128/MCB.23.1.349-358.2003
Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation
Abstract
RNA polymerase II (Pol II) Mediator plays an essential role in both basal and activated transcription. Previously, subunits of the Sin4 Mediator complex (Sin4, Pgd1, Gal11, and Med2) have been implicated in both positive and negative transcriptional regulation. Furthermore, it was proposed that this subcomplex constitutes an activator-binding domain. A yeast nuclear-extract system was used to investigate the biochemical role of the Sin4 complex. In contrast to previous findings, we found at least two general activator-independent roles for the Sin4 complex. First, mutations in sin4 and pgd1 destabilized the Pol II-Med complex, leading to a reduced rate and extent of preinitiation complex (PIC) formation both in the presence and absence of activators. Although reduced in amount compared with the wild type, PICs that are formed lacking the Sin4 complex are stable and can initiate transcription normally. Second, mutation of pgd1 causes partial disruption of the Sin4 complex and leads to a defect in transcription reinitiation. This defect is caused by dissociation of mutant Mediator from promoters after initiation, leading to nonfunctional Scaffold complexes. These results show that function of the Sin4 complex is not essential for transcription activation in a crude in vitro system but that it plays key roles in the general transcription mechanism.
Figures

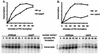





References
-
- Boube, M., L. Joulia, D. L. Cribbs, and H.-M. Bourbon. 2002. Evidence for a Mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. - PubMed
-
- Boyer, T. G., M. E. D. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. - PubMed
-
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
