A comparative cDNA microarray analysis reveals a spectrum of genes regulated by Pax6 in mouse lens
- PMID: 12485166
- PMCID: PMC2080869
- DOI: 10.1046/j.1365-2443.2002.00602.x
A comparative cDNA microarray analysis reveals a spectrum of genes regulated by Pax6 in mouse lens
Abstract
Background: Pax6 is a transcription factor that is required for induction, growth, and maintenance of the lens; however, few direct target genes of Pax6 are known.
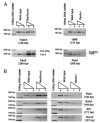
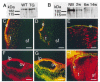
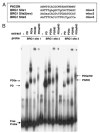
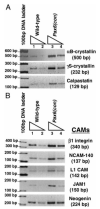
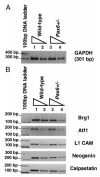
Results: In this report, we describe the results of a cDNA microarray analysis of lens transcripts from transgenic mice over-expressing Pax6 in lens fibre cells in order to narrow the field of potential direct Pax6 target genes. This study revealed that the transcript levels were significantly altered for 508 of the 9700 genes analysed, including five genes encoding the cell adhesion molecules beta1-integrin, JAM1, L1 CAM, NCAM-140 and neogenin. Notably, comparisons between the genes differentially expressed in Pax6 heterozygous and Pax6 over-expressing lenses identified 13 common genes, including paralemmin, GDIbeta, ATF1, Hrp12 and Brg1. Immunohistochemistry and Western blotting demonstrated that Brg1 is expressed in the embryonic and neonatal (2-week-old) but not in 14-week adult lenses, and confirmed altered expression in transgenic lenses over-expressing Pax6. Furthermore, EMSA demonstrated that the BRG1 promoter contains Pax6 binding sites, further supporting the proposition that it is directly regulated by Pax6.
Conclusions: These results provide a list of genes with possible roles in lens biology and cataracts that are directly or indirectly regulated by Pax6.
Figures


References
-
- Baulmann DC, Ohlmann A, Flugel-Koch C, Goswami S, Cvekl A, Tamm ER. Pax6 heterozygous eyes show defects in chamber angle differentiation that are associated with a wide spectrum of the other anterior eye segment abnormalities. Mech. Dev. 2002;118:3–17. - PubMed
-
- Belecky-Adams, Tomarev S, Li HS, et al. Pax-6, Prox 1, and Chx10 homeobox gene expression correlates with phenotypic fate of retinal precursor cells. Invest. Ophthalmol. Vis. Sci. 1997;38:1293–1303. - PubMed
-
- Bernier G, Vukovichn W, Neidhardt L, Herrmann BG, Gruss P. Isolation and characterization of a downstream target of Pax6 in the mammalian retinal primordium. Development. 2001;128:3987–3994. - PubMed
-
- Bultman S, Gebuhr T, Yee D, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6:1287–1295. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

