A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A
- PMID: 12485987
- PMCID: PMC139082
- DOI: 10.1093/emboj/cdf619
A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A
Abstract
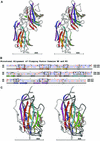
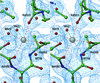
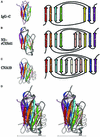
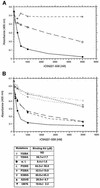
We report here the crystal structure of the minimal ligand-binding segment of the Staphylococcus aureus MSCRAMM, clumping factor A. This fibrinogen-binding segment contains two similarly folded domains. The fold observed is a new variant of the immunoglobulin motif that we have called DE-variant or the DEv-IgG fold. This subgroup includes the ligand-binding domain of the collagen-binding S.aureus MSCRAMM CNA, and many other structures previously classified as jelly rolls. Structure predictions suggest that the four fibrinogen-binding S.aureus MSCRAMMs identified so far would also contain the same DEv-IgG fold. A systematic docking search using the C-terminal region of the fibrinogen gamma-chain as a probe suggested that a hydrophobic pocket formed between the two DEv-IgG domains of the clumping factor as the ligand-binding site. Mutagenic substitution of residues Tyr256, Pro336, Tyr338 and Lys389 in the clumping factor, which are proposed to contact the terminal residues (408)AGDV(411) of the gamma-chain, resulted in proteins with no or markedly reduced affinity for fibrinogen.
Figures



References
-
- Blanchet C., Combet,C., Geourjon,C. and Deleage,G. (2000) MPSA: integrated system for multiple protein sequence analysis with client/server capabilities. Bioinformatics, 16, 286–287. - PubMed
-
- Bork P., Holm,L. and Sander,C. (1994) The immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set, which is close to that containing the variable domains. J. Mol. Biol., 242, 309–320. - PubMed
-
- Branden C. and Tooze,J. (1999) Introduction to Protein Structure, 2nd edn. Garland Publishing, New York, NY.
-
- Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

