HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA
- PMID: 12486004
- PMCID: PMC139084
- DOI: 10.1093/emboj/cdf663
HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA
Abstract
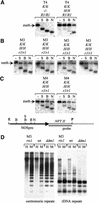
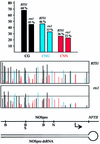
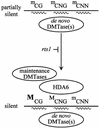
To analyze relationships between RNA signals, DNA methylation and chromatin modifications, we performed a genetic screen to recover Arabidopsis mutants defective in RNA-directed transcriptional silencing and methylation of a nopaline synthase promoter-neomycinphosphotransferase II (NOSpro- NPTII) target gene. Mutants were identified by screening for recovery of kanamycin resistance in the presence of an unlinked silencing complex encoding NOSpro double-stranded RNA. One mutant, rts1 (RNA-mediated transcriptional silencing), displayed moderate recovery of NPTII gene expression and partial loss of methylation in the target NOSpro, predominantly at symmetrical C(N)Gs. The RTS1 gene was isolated by positional cloning and found to encode a putative histone deacetylase, HDA6. The more substantial decrease in methylation of symmetrical compared with asymmetrical cytosines in rts1 mutants suggests that HDA6 is dispensable for RNA-directed de novo methylation, which results in intermediate methylation of cytosines in all sequence contexts, but is necessary for reinforcing primarily C(N)G methylation induced by RNA. Because CG methylation in centromeric and rDNA repeats was not reduced in rts1 mutants, HDA6 might be specialized for the RNA- directed pathway of genome modification.
Figures






References
-
- Amedeo P., Habu,Y., Afsar,K., Mittelsten Scheid,O. and Paszkowski,J. (2000) Disruption of the plant gene MOM releases transcriptional silencing of methylated transgenes. Nature, 405, 203–206. - PubMed
-
- Burgers W., Fuks,F. and Kouzarides,T. (2002) DNA methyltransferases get connected to chromatin. Trends Genet., 18, 275–277. - PubMed
-
- Cao X. and Jacobsen,S. (2002) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol., 12, 1138–1144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

