PNPase activity determines the efficiency of mRNA 3'-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts
- PMID: 12486011
- PMCID: PMC139106
- DOI: 10.1093/emboj/cdf686
PNPase activity determines the efficiency of mRNA 3'-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts
Abstract
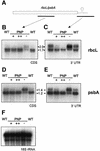
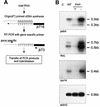
The exoribonuclease polynucleotide phosphorylase (PNPase) has been implicated in mRNA processing and degradation in bacteria as well as in chloroplasts of higher plants. Here, we report the first comprehensive in vivo study of chloroplast PNPase function. Modulation of PNPase activity in Arabidopsis chloroplasts by a reverse genetic approach revealed that, although this enzyme is essential for efficient 3'-end processing of mRNAs, it is insufficient to mediate transcript degradation. Surprisingly, we identified PNPase as also being indispensable for 3'-end maturation of 23S rRNA transcripts. Analysis of tRNA amounts in transgenic Arabidopsis plants suggests a direct correlation of PNPase activity and tRNA levels, indicating an additional function of this exoribo nuclease in tRNA decay. Moreover, the extent of polyadenylated mRNAs in chloroplasts is negatively correlated with PNPase activity. Together, our results attribute novel functions to PNPase in the metabolism of all major classes of plastid RNAs and suggest an unexpectedly complex role for PNPase in RNA processing and decay.
Figures





References
-
- Carpousis A.J., Vanzo,N.F. and Raynal,L.C. (1999) mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet., 15, 24–28. - PubMed
-
- Coburn G.A. and Mackie,G.A. (1999) Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol., 62, 55–108. - PubMed
-
- Decker C.J. (1998) The exosome: a versatile RNA processing machine. Curr. Biol., 8, R238–R240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

