Characterization of the 4-carboxy-4-hydroxy-2-oxoadipate aldolase gene and operon structure of the protocatechuate 4,5-cleavage pathway genes in Sphingomonas paucimobilis SYK-6
- PMID: 12486039
- PMCID: PMC141877
- DOI: 10.1128/JB.185.1.41-50.2003
Characterization of the 4-carboxy-4-hydroxy-2-oxoadipate aldolase gene and operon structure of the protocatechuate 4,5-cleavage pathway genes in Sphingomonas paucimobilis SYK-6
Abstract
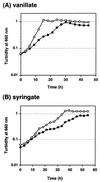
The protocatechuate (PCA) 4,5-cleavage pathway is the essential metabolic route for degradation of low-molecular-weight products derived from lignin by Sphingomonas paucimobilis SYK-6. In the 10.5-kb EcoRI fragment carrying the genes for PCA 4,5-dioxygenase (ligAB), 2-pyrone-4,6-dicarboxylate hydrolase (ligI), 4-oxalomesaconate hydratase (ligJ), and a part of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase (ligC), we found the ligK gene, which encodes 4-carboxy-4-hydroxy-2-oxoadipate (CHA) aldolase. The ligK gene was located 1,183 bp upstream of ligI and transcribed in the same direction as ligI. We also found the ligR gene encoding a LysR-type transcriptional activator, which was located 174 bp upstream of ligK. The ligK gene consists of a 684-bp open reading frame encoding a polypeptide with a molecular mass of 24,131 Da. The deduced amino acid sequence of ligK showed 57 to 88% identity with those of the corresponding genes recently reported in Sphingomonas sp. strain LB126, Comamonas testosteroni BR6020, Arthrobacter keyseri 12B, and Pseudomonas ochraceae NGJ1. The ligK gene was expressed in Escherichia coli, and the gene product (LigK) was purified to near homogeneity. Electrospray-ionization mass spectrometry indicated that LigK catalyzes not only the conversion of CHA to pyruvate and oxaloacetate but also that of oxaloacetate to pyruvate and CO(2). LigK is a hexamer, and its isoelectric point is 5.1. The K(m) for CHA and oxaloacetate are 11.2 and 136 micro M, respectively. Inactivation of ligK in S. paucimobilis SYK-6 resulted in the growth deficiency of vanillate and syringate, indicating that ligK encodes the essential CHA aldolase for catabolism of these compounds. Reverse transcription-PCR analysis revealed that the PCA 4,5-cleavage pathway genes of S. paucimobilis SYK-6 consisted of four transcriptional units, including the ligK-orf1-ligI-lsdA cluster, the ligJAB cluster, and the monocistronic ligR and ligC genes.
Figures




References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Davison, J., F. Brunel, A. Phanopoulos, D. Prozzi, and P. Terpstra. 1992. Cloning and sequencing of Pseudomonas genes determining sodium dodecyl sulfate biodegradation. Gene 114:19-24. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

