Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE
- PMID: 12486056
- PMCID: PMC141945
- DOI: 10.1128/JB.185.1.196-203.2003
Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE
Abstract
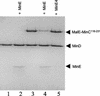
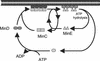
In Escherichia coli, the min system prevents division away from midcell through topological regulation of MinC, an inhibitor of Z-ring formation. The topological regulation involves oscillation of MinC between the poles of the cell under the direction of the MinDE oscillator. Since the mechanism of MinC involvement in the oscillation is unknown, we investigated the interaction of MinC with the other Min proteins. We observed that MinD dimerized in the presence of ATP and interacted with MinC. In the presence of a phospholipid bilayer, MinD bound to the bilayer and recruited MinC in an ATP-dependent manner. Addition of MinE to the MinCD-bilayer complex resulted in release of both MinC and MinD. The release of MinC did not require ATP hydrolysis, indicating that MinE could displace MinC from the MinD-bilayer complex. In contrast, MinC was unable to displace MinE bound to the MinD-bilayer complex. These results suggest that MinE induces a conformational change in MinD bound to the bilayer that results in the release of MinC. Also, it is argued that binding of MinD to the membrane activates MinC.
Figures







Similar articles
-
The bacterial cell division regulators MinD and MinC form polymers in the presence of nucleotide.FEBS Lett. 2015 Jan 16;589(2):201-6. doi: 10.1016/j.febslet.2014.11.047. Epub 2014 Dec 10. FEBS Lett. 2015. PMID: 25497011
-
ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro.J Bacteriol. 2003 Feb;185(3):735-49. doi: 10.1128/JB.185.3.735-749.2003. J Bacteriol. 2003. PMID: 12533449 Free PMC article.
-
Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE.Mol Microbiol. 1999 Oct;34(1):82-90. doi: 10.1046/j.1365-2958.1999.01575.x. Mol Microbiol. 1999. PMID: 10540287
-
Spatial control of the cell division site by the Min system in Escherichia coli.Environ Microbiol. 2013 Dec;15(12):3229-39. doi: 10.1111/1462-2920.12119. Epub 2013 Apr 9. Environ Microbiol. 2013. PMID: 23574354 Review.
-
MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation.Mol Microbiol. 2003 Apr;48(2):295-303. doi: 10.1046/j.1365-2958.2003.03427.x. Mol Microbiol. 2003. PMID: 12675792 Review.
Cited by
-
Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes.J Bacteriol. 2005 Mar;187(6):2163-74. doi: 10.1128/JB.187.6.2163-2174.2005. J Bacteriol. 2005. PMID: 15743965 Free PMC article.
-
Analysis of MinD mutations reveals residues required for MinE stimulation of the MinD ATPase and residues required for MinC interaction.J Bacteriol. 2005 Jan;187(2):629-38. doi: 10.1128/JB.187.2.629-638.2005. J Bacteriol. 2005. PMID: 15629934 Free PMC article.
-
Self-assembly of MinE on the membrane underlies formation of the MinE ring to sustain function of the Escherichia coli Min system.J Biol Chem. 2014 Aug 1;289(31):21252-66. doi: 10.1074/jbc.M114.571976. Epub 2014 Jun 9. J Biol Chem. 2014. PMID: 24914211 Free PMC article.
-
Characterization of C-terminal structure of MinC and its implication in evolution of bacterial cell division.Sci Rep. 2017 Aug 8;7(1):7627. doi: 10.1038/s41598-017-08213-5. Sci Rep. 2017. PMID: 28790446 Free PMC article.
-
Role of signature lysines in the deviant walker a motifs of the ArsA ATPase.Biochemistry. 2010 Jan 19;49(2):356-64. doi: 10.1021/bi901681v. Biochemistry. 2010. PMID: 20000479 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

