Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1
- PMID: 12486058
- PMCID: PMC141832
- DOI: 10.1128/JB.185.1.210-220.2003
Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1
Abstract
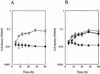
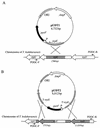
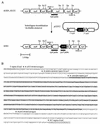
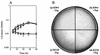
In contrast to the high accumulation in sequence data for hyperthermophilic archaea, methodology for genetically manipulating these strains is still at an early stage. This study aimed to develop a gene disruption system for the hyperthermophilic euryarchaeon Thermococcus kodakaraensis KOD1. Uracil-auxotrophic mutants with mutations in the orotidine-5'-monophosphate decarboxylase gene (pyrF) were isolated by positive selection using 5-fluoroorotic acid (5-FOA) and used as hosts for further transformation experiments. We then attempted targeted disruption of the trpE locus in the host strain by homologous recombination, as disruption of trpE was expected to result in tryptophan auxotrophy, an easily detectable phenotype. A disruption vector harboring the pyrF marker within trpE was constructed for double-crossover recombination. The host cells were transformed with the exogenous DNA using the CaCl(2) method, and several transformants could be selected based on genetic complementation. Genotypic and phenotypic analyses of a transformant revealed the unique occurrence of targeted disruption, as well as a phenotypic change of auxotrophy from uracil to tryptophan caused by integration of the wild-type pyrF into the host chromosome at trpE. As with the circular plasmid, gene disruption with linear DNA was also possible when the homologous regions were relatively long. Shortening these regions led to predominant recombination between the pyrF marker in the exogenous DNA and the mutated allele on the host chromosome. In contrast, we could not obtain trpE disruptants by insertional inactivation using a vector designed for single-crossover recombination. The gene targeting system developed in this study provides a long-needed tool in the research on hyperthermophilic archaea and will open the way to a systematic, genetic approach for the elucidation of unknown gene function in these organisms.
Figures


Similar articles
-
Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis.Appl Environ Microbiol. 2005 Jul;71(7):3889-99. doi: 10.1128/AEM.71.7.3889-3899.2005. Appl Environ Microbiol. 2005. PMID: 16000802 Free PMC article.
-
Development of a versatile method for targeted gene deletion and insertion by using the pyrF gene in the psychrotrophic bacterium, Shewanella livingstonensis Ac10.J Biosci Bioeng. 2016 Dec;122(6):645-651. doi: 10.1016/j.jbiosc.2016.06.004. Epub 2016 Jul 9. J Biosci Bioeng. 2016. PMID: 27401770
-
Genetic manipulations of the hyperthermophilic piezophilic archaeon Thermococcus barophilus.Appl Environ Microbiol. 2014 Apr;80(7):2299-306. doi: 10.1128/AEM.00084-14. Epub 2014 Jan 31. Appl Environ Microbiol. 2014. PMID: 24487541 Free PMC article.
-
An overview of 25 years of research on Thermococcus kodakarensis, a genetically versatile model organism for archaeal research.Folia Microbiol (Praha). 2020 Feb;65(1):67-78. doi: 10.1007/s12223-019-00730-2. Epub 2019 Jul 8. Folia Microbiol (Praha). 2020. PMID: 31286382 Review.
-
Sequential gene deletions in Hypocrea jecorina using a single blaster cassette.Curr Genet. 2005 Sep;48(3):204-11. doi: 10.1007/s00294-005-0011-8. Epub 2005 Oct 12. Curr Genet. 2005. PMID: 16091959 Review.
Cited by
-
Progress and Challenges in Archaeal Genetic Manipulation.Methods Mol Biol. 2022;2522:25-31. doi: 10.1007/978-1-0716-2445-6_2. Methods Mol Biol. 2022. PMID: 36125741
-
Progress and Challenges in Archaeal Cell Biology.Methods Mol Biol. 2022;2522:365-371. doi: 10.1007/978-1-0716-2445-6_24. Methods Mol Biol. 2022. PMID: 36125763
-
Development of an Effective 6-Methylpurine Counterselection Marker for Genetic Manipulation in Thermococcus barophilus.Genes (Basel). 2018 Feb 7;9(2):77. doi: 10.3390/genes9020077. Genes (Basel). 2018. PMID: 29414865 Free PMC article.
-
Chromosomal domain formation by archaeal SMC, a roadblock protein, and DNA structure.Nat Commun. 2025 Feb 19;16(1):1312. doi: 10.1038/s41467-025-56197-y. Nat Commun. 2025. PMID: 39971902 Free PMC article.
-
The archaeal Ced system imports DNA.Proc Natl Acad Sci U S A. 2016 Mar 1;113(9):2496-501. doi: 10.1073/pnas.1513740113. Epub 2016 Feb 16. Proc Natl Acad Sci U S A. 2016. PMID: 26884154 Free PMC article.
References
-
- Aagaard, C., I. Leviev, R. N. Aravalli, P. Forterre, D. Prieur, and R. A. Garrett. 1996. General vectors for archaeal hyperthermophiles: strategies based on a mobile intron and a plasmid. FEMS Microbiol. Rev. 18:93-104. - PubMed
-
- Adams, M. W., and R. M. Kelly. 1998. Finding and using hyperthermophilic enzymes. Trends Biotechnol. 16:329-332. - PubMed
-
- Aravalli, R. N., and R. A. Garrett. 1997. Shuttle vectors for hyperthermophilic archaea. Extremophiles 1:183-191. - PubMed
-
- Beckwith, J. R., A. B. Pardee, R. Austrian, and F. Jacob. 1962. Coordination of the synthesis of the enzymes in the pyrimidine pathway of E. coli. J. Mol. Biol. 5:618-634. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

