Multiple promoter inversions generate surface antigenic variation in Mycoplasma penetrans
- PMID: 12486060
- PMCID: PMC141813
- DOI: 10.1128/JB.185.1.231-242.2003
Multiple promoter inversions generate surface antigenic variation in Mycoplasma penetrans
Abstract
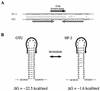
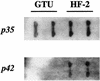
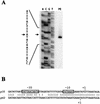
Mycoplasma penetrans is a newly identified species of the genus MYCOPLASMA: It was first isolated from a urine sample from a human immunodeficiency virus (HIV)-infected patient. M. penetrans changes its surface antigen profile with high frequency. The changes originate from ON<==>OFF phase variations of the P35 family of surface membrane lipoproteins. The P35 family lipoproteins are major antigens recognized by the human immune system during M. penetrans infection and are encoded by the mpl genes. Phase variations of P35 family lipoproteins occur at the transcriptional level of mpl genes; however, the precise genetic mechanisms are unknown. In this study, the molecular mechanisms of surface antigen profile change in M. penetrans were investigated. The focus was on the 46-kDa protein that is present in M. penetrans strain HF-2 but not in the type strain, GTU. The 46-kDa protein was the product of a previously reported mpl gene, pepIMP13, with an amino-terminal sequence identical to that of the P35 family lipoproteins. Nucleotide sequencing analysis of the pepIMP13 gene region revealed that the promoter-containing 135-bp DNA of this gene had the structure of an invertible element that functioned as a switch for gene expression. In addition, all of the mpl genes of M. penetrans HF-2 were identified using the whole-genome sequence data that has recently become available for this bacterium. There are at least 38 mpl genes in the M. penetrans HF-2 genome. Interestingly, most of these mpl genes possess invertible promoter-like sequences, similar to those of the pepIMP13 gene promoter. A model for the generation of surface antigenic variation by multiple promoter inversions is proposed.
Figures






References
-
- Bhugra, B., L. L. Voelker, N. Zou, H. Yu, and K. Dybvig. 1995. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol. Microbiol. 18:703-714. - PubMed
-
- Chambaud, I., H. Wroblewski, and A. Blanchard. 1999. Interactions between mycoplasma lipoproteins and the host immune system. Trends Microbiol. 7:493-499. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

