The crystal structure of the phosphorylation domain in PhoP reveals a functional tandem association mediated by an asymmetric interface
- PMID: 12486062
- PMCID: PMC141828
- DOI: 10.1128/JB.185.1.254-261.2003
The crystal structure of the phosphorylation domain in PhoP reveals a functional tandem association mediated by an asymmetric interface
Abstract
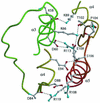
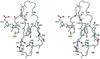
PhoP from Bacillus subtilis belongs to the OmpR subfamily of response regulators. It regulates the transcription of several operons and participates in a signal transduction network that controls adaptation of the bacteria to phosphate deficiency. The receiver domains of two members of this subfamily, PhoB from Escherichia coli and DrrD from Thermotoga maritima, have been structurally characterized. These modules have similar overall folds but display remarkable differences in the conformation of the beta4-alpha4 and alpha4 regions. The crystal structure of the receiver domain of PhoP (PhoPN) described in this paper illustrates yet another geometry in this region. Another major issue of the structure determination is the dimeric state of the protein and the novel mode of association between receiver domains. The protein-protein interface is provided by two different surfaces from each protomer, and the tandem unit formed through this asymmetric interface leaves free interaction surfaces. This design is well suited for further association of PhoP dimers to form oligomeric structures. The interprotein interface buries 970 A(2) from solvent and mostly involves interactions between charged residues. As described in the accompanying paper, mutations of a single residue in one salt bridge shielded from solvent prevented dimerization of the unphosphorylated and phosphorylated response regulator and had drastic functional consequences. The three structurally documented members of the OmpR family (PhoB, DrrD, and PhoP) provide a framework to consider possible relationships between structural features and sequence signatures in critical regions of the receiver domains.
Figures
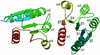

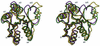

References
-
- Baikalov, I., I. Schroder, M. Kaczor-Grzeskowiak, K. Grzeskowiak, R. P. Gunsalus, and R. E. Dickerson. 1996. Structure of the Escherichia coli response regulator NarL. Biochemistry 35:11053-11061. - PubMed
-
- Birck, C., L. Mourey, P. Gouet, B. Fabry, J. Schumacher, P. Rousseau, D. Kahn, and J. P. Samama. 1999. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure 7:1505-1515. - PubMed
-
- Blanco, A. G., M. Sola, F. X. Gomis-Rüth, and M. Coll. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10:701-713. - PubMed
-
- Bourret, R. B., N. W. Charon, A. M. Stock, and A. H. West. 2002. Bright lights, abundant operons—fluorescence and genomic technologies advance studies of bacterial locomotion and signal transduction: review of the BLAST meeting, Cuernavaca, Mexico, 14 to 19 January 2001 J. Bacteriol. 184:1-17. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

