Evolutionary history of hrgA, which replaces the restriction gene hpyIIIR in the hpyIII locus of Helicobacter pylori
- PMID: 12486066
- PMCID: PMC141823
- DOI: 10.1128/JB.185.1.295-301.2003
Evolutionary history of hrgA, which replaces the restriction gene hpyIIIR in the hpyIII locus of Helicobacter pylori
Erratum in
- J Bacteriol. 2003 Mar;185(6):2059
Abstract
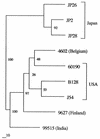
A recently identified Helicobacter pylori gene, hrgA, was previously reported to be present in 70 (33%) of 208 strains examined (T. Ando, T. M. Wassenaar, R. M. Peek, R. A. Aras, A. I. Tschumi, L.-J. Van Doorn, K. Kusugami, and M. J. Blaser, Cancer Res. 62:2385-2389, 2002). Sequence analysis of nine such strains indicated that in each strain hrgA replaced hpyIIIR, which encodes a restriction endonuclease and which, together with the gene for its cognate methyltransferase, constitutes the hpyIII locus. As a consequence of either the hrgA insertion or independent mutations, hpyIIIM function was lost in 11 (5%) of the 208 strains examined, rendering chromosomal DNA sensitive to MboI digestion. The evolutionary history of the locus containing either hpyIII or hrgA was reconstructed. By homologous recombination involving flanking sequences, hrgA and hpyIIIR can replace one another in the hpyIII locus, and there is simultaneous replacement of several flanking genes. These findings, combined with the hpyIM/iceA2 locus discovered previously, suggest that the two most strongly conserved methylase genes of H. pylori, hpyIIIM and hpyIM, are both preceded by alternative genes that compete for presence at their loci.
Figures


Similar articles
-
The Helicobacter pylori restriction endonuclease-replacing gene, hrgA, and clinical outcome: comparison of East Asia and Western countries.Dig Dis Sci. 2004 Sep;49(9):1551-5. doi: 10.1023/b:ddas.0000042263.18541.ec. Dig Dis Sci. 2004. PMID: 15481336
-
A Helicobacter pylori restriction endonuclease-replacing gene, hrgA, is associated with gastric cancer in Asian strains.Cancer Res. 2002 Apr 15;62(8):2385-9. Cancer Res. 2002. PMID: 11956101
-
Association of Helicobacter pylori restriction endonuclease-replacing gene, hrgA with overt gastrointestinal diseases.Arq Gastroenterol. 2008 Jul-Sep;45(3):225-9. doi: 10.1590/s0004-28032008000300011. Arq Gastroenterol. 2008. PMID: 18852951
-
Restriction-modification systems may be associated with Helicobacter pylori virulence.J Gastroenterol Hepatol. 2010 May;25 Suppl 1:S95-8. doi: 10.1111/j.1440-1746.2009.06211.x. J Gastroenterol Hepatol. 2010. PMID: 20586875 Review.
-
Contributions of genome sequencing to understanding the biology of Helicobacter pylori.Annu Rev Microbiol. 1999;53:353-87. doi: 10.1146/annurev.micro.53.1.353. Annu Rev Microbiol. 1999. PMID: 10547695 Review.
Cited by
-
Identification of new homologs of PD-(D/E)XK nucleases by support vector machines trained on data derived from profile-profile alignments.Nucleic Acids Res. 2011 Mar;39(4):1187-96. doi: 10.1093/nar/gkq958. Epub 2010 Oct 20. Nucleic Acids Res. 2011. PMID: 20961958 Free PMC article.
-
Prevalence, Virulence Genes, Phylogenetic Analysis, and Antimicrobial Resistance Profile of Helicobacter Species in Chicken Meat and Their Associated Environment at Retail Shops in Egypt.Foods. 2022 Jun 26;11(13):1890. doi: 10.3390/foods11131890. Foods. 2022. PMID: 35804706 Free PMC article.
-
The Helicobacter pylori HpyAXII restriction-modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components.Nucleic Acids Res. 2008 Dec;36(21):6893-906. doi: 10.1093/nar/gkn718. Epub 2008 Oct 31. Nucleic Acids Res. 2008. PMID: 18978016 Free PMC article.
-
The Helicobacter pylori restriction endonuclease-replacing gene, hrgA, and clinical outcome: comparison of East Asia and Western countries.Dig Dis Sci. 2004 Sep;49(9):1551-5. doi: 10.1023/b:ddas.0000042263.18541.ec. Dig Dis Sci. 2004. PMID: 15481336
-
High frequency of gastric colonization with multiple Helicobacter pylori strains in Venezuelan subjects.J Clin Microbiol. 2005 Jun;43(6):2635-41. doi: 10.1128/JCM.43.6.2635-2641.2005. J Clin Microbiol. 2005. PMID: 15956377 Free PMC article.
References
-
- Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. - PubMed
-
- Alm, R. A., and T. J. Trust. 1999. Analysis of the genetic diversity of Helicobacter pylori: the role of two genomes. J. Mol. Med. 77:834-846. - PubMed
-
- Ando, T., T. M. Wassenaar, R. M. Peek, R. A. Aras, A. I. Tschumi, L.-J. Van Doorn, K. Kusugami, and M. J. Blaser. 2002. A Helicobacter pylori restriction endonuclease replacing gene, hrgA, associated with gastric cancer in Asian strains. Cancer Res. 62:2385-2389. - PubMed
-
- Bennett, S. P., and S. E. Halford. 1989. Recognition of DNA by type II restriction enzymes. Curr. Top. Cell. Regul. 30:57-104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

