Membrane topology of the ZntB efflux system of Salmonella enterica serovar Typhimurium
- PMID: 12486076
- PMCID: PMC141924
- DOI: 10.1128/JB.185.1.374-376.2003
Membrane topology of the ZntB efflux system of Salmonella enterica serovar Typhimurium
Abstract
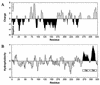
The membrane topology of the ZntB Zn(2+) transport protein of Salmonella enterica serovar Typhimurium was determined by constructing deletion derivatives of the protein and genetically fusing them to blaM or lacZ cassettes. The enzymatic activities of the hybrid proteins indicate that ZntB is a bitopic integral membrane protein consisting largely of two independent domains. The first 266 amino acids form a large, highly charged domain within the cytoplasm, while the remaining 61 residues form a small membrane domain containing two membrane-spanning segments. The overall orientation towards the cytoplasm is consistent with the ability of ZntB to facilitate zinc efflux.
Figures
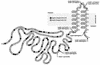
Similar articles
-
Sequence and topology of the CorA magnesium transport systems of Salmonella typhimurium and Escherichia coli. Identification of a new class of transport protein.J Biol Chem. 1993 Jul 5;268(19):14071-80. J Biol Chem. 1993. PMID: 8314774
-
ZntB is a novel Zn2+ transporter in Salmonella enterica serovar Typhimurium.J Bacteriol. 2002 Aug;184(16):4369-73. doi: 10.1128/JB.184.16.4369-4373.2002. J Bacteriol. 2002. PMID: 12142406 Free PMC article.
-
X-ray crystallography and isothermal titration calorimetry studies of the Salmonella zinc transporter ZntB.Structure. 2011 May 11;19(5):700-10. doi: 10.1016/j.str.2011.02.011. Structure. 2011. PMID: 21565704 Free PMC article.
-
Membrane topology of the Salmonella enterica serovar Typhimurium Group B O-antigen translocase Wzx.FEMS Microbiol Lett. 2008 Oct;287(1):76-84. doi: 10.1111/j.1574-6968.2008.01295.x. Epub 2008 Aug 14. FEMS Microbiol Lett. 2008. PMID: 18707624
-
The family of organo-phosphate transport proteins includes a transmembrane regulatory protein.J Bioenerg Biomembr. 1993 Dec;25(6):637-45. doi: 10.1007/BF00770251. J Bioenerg Biomembr. 1993. PMID: 8144492 Review.
Cited by
-
Transport of magnesium and other divalent cations: evolution of the 2-TM-GxN proteins in the MIT superfamily.Mol Genet Genomics. 2005 Oct;274(3):205-16. doi: 10.1007/s00438-005-0011-x. Epub 2005 Oct 20. Mol Genet Genomics. 2005. PMID: 16179994
-
Membrane topology analysis of cyclic glucan synthase, a virulence determinant of Brucella abortus.J Bacteriol. 2004 Nov;186(21):7205-13. doi: 10.1128/JB.186.21.7205-7213.2004. J Bacteriol. 2004. PMID: 15489431 Free PMC article.
-
Resistance to Metals Used in Agricultural Production.Microbiol Spectr. 2018 Apr;6(2):10.1128/microbiolspec.arba-0025-2017. doi: 10.1128/microbiolspec.ARBA-0025-2017. Microbiol Spectr. 2018. PMID: 29676247 Free PMC article. Review.
-
The unique nature of mg2+ channels.Physiology (Bethesda). 2008 Oct;23:275-85. doi: 10.1152/physiol.00019.2008. Physiology (Bethesda). 2008. PMID: 18927203 Free PMC article. Review.
-
E. coli allantoinase is activated by the downstream metabolic enzyme, glycerate kinase, and stabilizes the putative allantoin transporter by direct binding.Sci Rep. 2023 May 5;13(1):7345. doi: 10.1038/s41598-023-31812-4. Sci Rep. 2023. PMID: 37147430 Free PMC article.
References
-
- Argos, P., and J. K. Rao. 1986. Prediction of protein structure. Methods Enzymol. 130:185-207. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
-
- Broome-Smith, J. K., M. Tadayyon, and Y. Zhang. 1990. Beta-lactamase as a probe of membrane protein assembly and protein export. Mol. Microbiol. 4:1637-1644. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

